


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 16-12-2015
Date: 24-12-2015
Date: 3-5-2021
|
Mutation Detection
Since the early days of gene product analysis, electrophoresis-based methods have prevailed as the most common means for mutation detection. With the completion of the Human Genome Project and the production of the reference DNA sequence of the genome, there has been an increasing demand for faster and improved detection methods of polymorphic markers in the effort to map and to establish the function of all genes.
Although the role of classical DNA sequencing has remained a prominent weapon in the armoury for SNP detection,56 a variety of alternative laboratory approaches (Table ) have been developed to screen for, and type, new and existing SNPs. These methods include single-strand conformation polymorphism (SSCP), denaturing high-performance
Table :Semi-quantitative comparison of some methods for discovering new SNPs (detection) and for typing known SNPs (screening) in PCR products
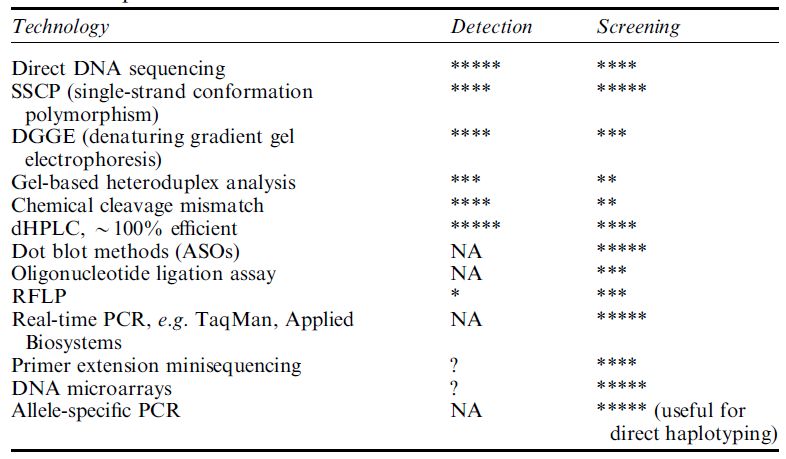
liquid chromatography (dHPLC) and microarray technology, which perhaps offers the greatest promise for the future.
1- Single-stranded Conformation Polymorphism (SSCP)
Originally described by Orita et al. in 1989,58 SSCP has been widely applied in the field of human genetics, where it has been used both for SNP detection and for SNP discovery,59 although it is generally preferred for the former application. The essence is to denature PCR products to make them single stranded and then to separate them by gel electrophoresis under non-denaturing conditions. In the absence of strong denaturants, a single-stranded fragment of DNA will adopt a unique and specific three-dimensional conformation as it attempts to fold into the most stable structure. The mutated form will adopt a different conformation to its wild-type counterpart. The differences in the conformers can be assessed on electrophoresis gels, where a heterozygous sample normally displays four bands, one for each denatured strand, and homozygotes normally display two bands (Figure 1). For mutation detection, the resolving power of the technique is improved by scanning PCR fragments that are less than 300 base pairs in length. Provided that PCR-SSCP analysis is conducted under appropriate conditions, for example by repeating each electrophoresis experiment at two running temperatures or by varying the amount of mild denaturant included in the gel (such as formamide), it has been shown to be an efficient approach to discovering new mutations. Initially SSCPs were detected using autoradiography of radiolabelled PCR products, followed later by silver staining to visualise unlabelled DNA fragments. A semi-automated method, PLACE-SSCP, has been developed in which the products of the PCR are labelled with fluorescent dyes and analysed by capillary electrophoresis under SSCP conditions. As with many other mutation detection techniques, SSCP analysis is amenable tomul tiplexed PCR formats where several PCR products labelled with different fluorophores are analysed in the same electrophoretic lane.

Figure 1 Principle of SSCP analysis. In the presence of heat and formamide, the dsDNA PCR products from a heterozygote are denatured for several minutes to form single-stranded DNA (ssDNA). Immediately before loading on to a non-denaturing electrophoresis gel, the samples are cooled on ice to encourage the formation of ssDNA conformers. Each conformer has a unique electrophoretic mobility as shown on the gel diagram.
2- Denaturing High-performance Liquid Chromatography(DHPLC)
DHPLC has been shown to be a fast and reliable method for SNP detection and discovery. The technique is based on the analysis of homoduplexes and heteroduplexes formed between reference and mutant DNA molecules by iron-pair reversed-phase high-performance liquid chromatography under partially denaturing conditions. For DNA fragments of between about 100 and 1500 base pairs, DHPLC has been shown to be capable of detecting all single base substitutions and also
small insertions and deletions.
To perform the analysis, DNA fragments amplified by PCR from reference and test chromosomes are mixed together, fully denatured at 95 °C and allowed to re-anneal by reducing the temperature slowly to about 25 °C. This results in three classes of duplexes: homoduplexes of the reference DNA and mutant DNAs and, third, mismatched heteroduplexes formed between reference and mutant DNA. Mismatched
heteroduplexes and homoduplexes are then introduced to an ion pair of the reversed-phase HPLC flow path where they bind to the HPLC capillary. The temperature is then increased to a threshold level, usually between 50 and 60 °C, whereupon the DNA duplexes become partially denatured. Initially the duplexes are retained in the HPLC capillary through ionic binding interactions between the negatively charged phosphate backbone of the partially denatured DNA fragments and the beads in the cartridge, which are coated with the positively charged ionpair reagent TEAA. The column is then eluted with a TEAA–acetonitrile gradient under conditions such that the heteroduplexes with a mismatched base pair(s) elute before the more stable homoduplexes.
Note that for SNP detection purposes, the two homoduplexes are indistinguishable. The eluted fragments then pass through a UV detector and the absorbance is measured and the data are analysed by computer. Two sets of peaks are observed for each duplex (Figure 2). The method is rapid, taking on average 7 min to analyse a genotype, and amenable to multiplexing through the use of fluorescence primer detection analogous to high-throughput DNA sequencing strategies. From a practical perspective, the method requires the use of Pfu polymerase instead of Taq polymerase for generating PCR products. Pfu has proof-reading properties which minimise the introduction of confusing PCR induced mutations.
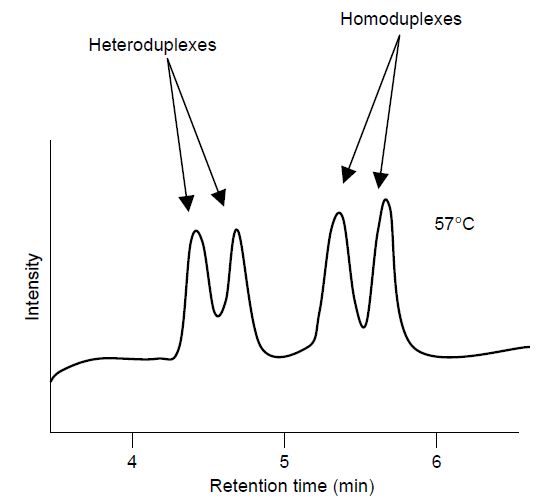
Figure 2 Schematic of dHPLC analysis of PCR products showing that the homoduplex and heteroduplex DNA species are differentially eluted when the column is held at the Tm of the PCR fragment, in this case 57 °C. N.B. to detect homozygous point mutations, the mutant PCR product is mixed with reference wild-type DNA.
3- DNA Microarrays
The need for rapid, high-quality, comprehensive gene-related data for a variety of purposes has stimulated the development of DNA microarrays or biochips. These provide a radically different approach to largescale characterisation of genes and gene expression. DNA microarrays are a departure from electrophoresis-based approaches and currently represent the extreme end of miniaturisation. A microarray is an ordered array of many thousands of DNA oligonucleotides, either singlestranded oligonucleotides or double-stranded cDNAs, attached (either by direct printing or in situ synthesis of short oligonucleotides) to a glass or silicon ‘chip’ that is about the size of a microscope coverslip. The bound DNA can then be hybridised with test DNA or RNA which has been labelled with one or two fluorescent dyes depending on the experimental design. Following an incubation step, the unhybridised material is washed away and the result is recorded using a confocal laser scanner. The data are collected and displayed automatically using dedicated computer programs.
Initially microarrays were designed to measure mRNA transcripts from thousands of genes in a single experiment. This enabled the physiological state of cells and overall gene expression pattern to be correlated. For example, transcriptional profiles have been obtained for many types of human cancer and the accumulated data promise to lead the way to a better understanding of neoplasia and new therapeutic targets.
DNA microarrays are not limited to gene expression studies. The first genotyping biochips were devised to identify key mutations in highly variable medically important genes and genomes such as the tumour suppressor gene TP53 (OMIM 191170)73 and the human immunodeficiency virus (HIV). In both cases, successful genotyping of clinical material is achieved using the microarray approach. More recently, genotyping chips capable of assigning SNP alleles on a ‘whole genome’ basis have been devised, which have applications in a variety of post-genomic projects such as the HapMap initiative to map all human variations in different population groups. Three commercially available microarrays are available that make it possible to analyse simultaneously many thousands of SNPs in an individual’s DNA. Two microarrays from Affymetrix, with roughly 100 000 and 500 000 SNPs, respectively, and a microarray from Illumina, with more than 300 000 SNPs, enable a large sample of the genetic variation of an individual to be assessed in a single experiment. Such tools should improve the opportunities for correlating common diseases with genetic differences in statistically high-powered association studies. Commercially available microarrays have now been developed to monitor SNP profiles in economically or scientifically important non-humans such as cattle and mice. However, the current estimate is that there are more than 10 million SNP sites in the human genome and considerable future developments will probably be required to give the required coverage of SNPs in all human populations sufficient for genome-wide association studies.



|
|
|
|
تفوقت في الاختبار على الجميع.. فاكهة "خارقة" في عالم التغذية
|
|
|
|
|
|
|
أمين عام أوبك: النفط الخام والغاز الطبيعي "هبة من الله"
|
|
|
|
|
|
|
المجمع العلمي ينظّم ندوة حوارية حول مفهوم العولمة الرقمية في بابل
|
|
|