


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 2025-02-27
Date: 4-11-2020
Date: 1-6-2021
|
16S rRNA Plays an Active Role in Translation
KEY CONCEPT
- 16S rRNA plays an active role in the functions of the 30S subunit. It directly interacts with mRNA, the 50S subunit, and the anticodons of tRNAs in the P and A sites.
The ribosome was originally viewed as a collection of proteins with various catalytic activities held together by protein–protein interactions and RNA–protein interactions. However, the discovery of RNA molecules with catalytic activities immediately suggests that rRNA might play a more active role in ribosome function. Evidence now suggests that rRNA interacts with mRNA or tRNA at each stage of translation and that the proteins are necessary to maintain the rRNA in a structure in which it can perform the catalytic functions. Several interactions involve specific regions of rRNA:
- The 3′ terminus of the 16S rRNA interacts directly with mRNA at initiation.
- Specific regions of 16S rRNA interact directly with the anticodon regions of tRNAs in both the A site and the P site. Similarly, 23S rRNA interacts with the CCA terminus of peptidyl-tRNA in both the P site and A site.
- Subunit interaction involves interactions between 16S and 23S rRNAs (see the section earlier in this chapter titled Ribosomal RNA Is Found Throughout Both Ribosomal Subunits).
A lot of information about the individual steps of bacterial translation has been obtained by using antibiotics that inhibit the process at particular stages. The target for the antibiotic can be identified by the component in which resistant mutations occur. Some antibiotics act on individual ribosomal proteins, but several act on rRNA, which suggests that the rRNA is involved with many or even all of the functions of the ribosome.
Two types of approaches have been used to investigate the functions of rRNA. Structural studies show that particular regions of rRNA are located in important sites of the ribosome and that chemical modifications of these bases impede particular ribosomal functions. In addition, mutations identify nucleotides in rRNA that are required for particular ribosomal functions. Figure 1 summarizes the sites in 16S rRNA that have been identified by these means.
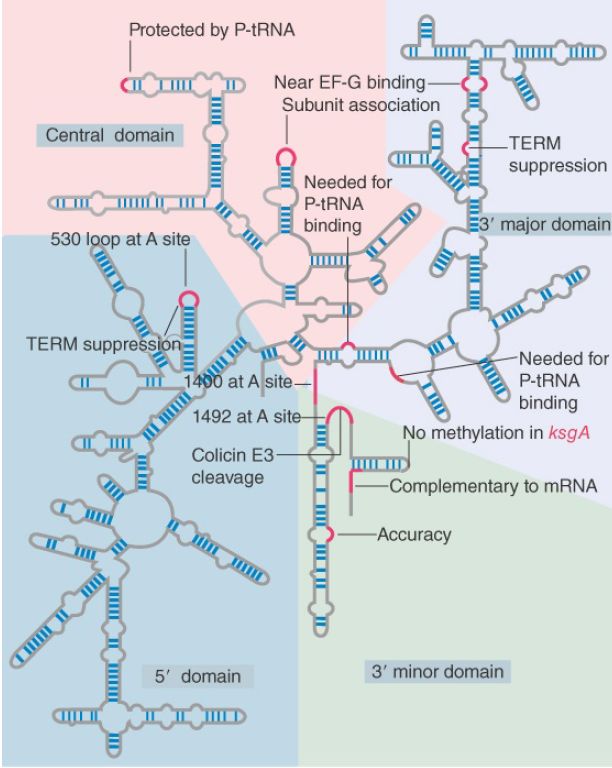
FIGURE 1. Some sites in 16S rRNA are protected from chemical probes when 50S subunits join 30S subunits or when aminoacyl-tRNA binds to the A site. Others are the sites of mutations that affect translation. TEM suppression sites may affect termination at some or several termination codons. The large colored blocks indicate the four domains of the rRNA.
An indication of the importance of the 3′ end of 16S rRNA is given by its susceptibility to the lethal agent colicin E3. Produced by some bacteria, colicin cleaves about 50 nucleotides from the 3′ end of the 16S rRNA of E. coli. The cleavage entirely abolishes initiation of translation. The region that is cleaved has several important functions: binding the factor IF-3, recognition of mRNA, and binding of tRNA.
The 3′ end of the 16S rRNA is directly involved in the initiation reaction by pairing with the Shine–Dalgarno sequence in the ribosome-binding site of mRNA. Another direct role for the 3′ end of 16S rRNA in translation is shown by the properties of kasugamycinresistant mutants, which lack certain modifications in 16S rRNA.
Kasugamycin blocks initiation of translation. Resistant mutants (called ksgA) lack a methylase enzyme that introduces four methyl groups into two adjacent adenines at a site near the 3′ terminus of the 16S rRNA. The methylation generates the highly conserved sequence G–m26 A–m26A, which is found in both prokaryotic and eukaryotic small rRNAs. The methylated sequence is involved in the joining of the 30S and 50S subunits, which, in turn, is connected also with the retention of initiator tRNA in the complete ribosome. Kasugamycin causes fMet-tRNAf to be released from the sensitive (methylated) ribosomes, but the resistant ribosomes are able to retain the initiator.
Changes in the structure of 16S rRNA occur when ribosomes are engaged in translation, as seen by protection of particular bases against chemical attack. The individual sites fall into a few groups that are concentrated in the 3′ minor and central domains. Although the locations are dispersed in the linear sequence of 16S rRNA, it seems likely that base positions involved in the same function are actually close together in the tertiary structure.
Some of the changes in 16S rRNA are triggered by joining with 50S subunits, binding of mRNA, or binding of tRNA. They indicate that these events are associated with changes in ribosome conformation that affect the exposure of rRNA. They do not necessarily indicate direct participation of rRNA in these functions. One change that occurs during translation is shown in Figure 2; it involves a local movement to change the nature of a short duplex sequence.
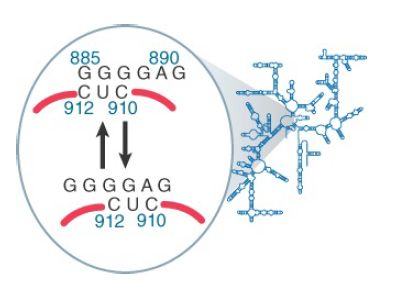
FIGURE 2. A change in conformation of 16S rRNA may occur during translation.
The 16S rRNA is involved in both A site and P site function, and significant changes in its structure occur when these sites are occupied. Certain distinct regions are protected by tRNA bound in the A site. One is the 530 loop (which also is the site of a mutation that prevents termination at the UAA, UAG, and UGA codons). The other is the 1400 to 1500 region (so called because bases 1399 to 1492 and the adenines at 1492 and 1493 are two single-stranded stretches that are connected by a long hairpin). All of the effects that tRNA binding has on 16S rRNA can be produced by the isolated oligonucleotide of the anticodon stem-loop, thus tRNA–30S subunit binding must involve this region.
The adenines at 1492 and 1493 provide a mechanism for detecting properly paired codon–anticodon complexes. The principle of the interaction is that the structure of the 16S rRNA responds to the structure of the first two base pairs in the minor groove of the duplex formed by the codon–anticodon interaction. Modification of the N1 position of either base 1492 or 1493 in rRNA prevents tRNA from binding in the A site. However, mutations at 1492 or 1493 can be suppressed by the introduction of fluorine at the 2′ position of the corresponding bases in mRNA (which restores the interaction).
Figure 3 shows that codon–anticodon pairing allows the N1 of each adenine to interact with the 2′–OH in the mRNA backbone. The interaction stabilizes the association of tRNA with the A site. When an incorrect tRNA enters the A site, the structure of the codon–anticodon complex is distorted, and this interaction cannot occur.
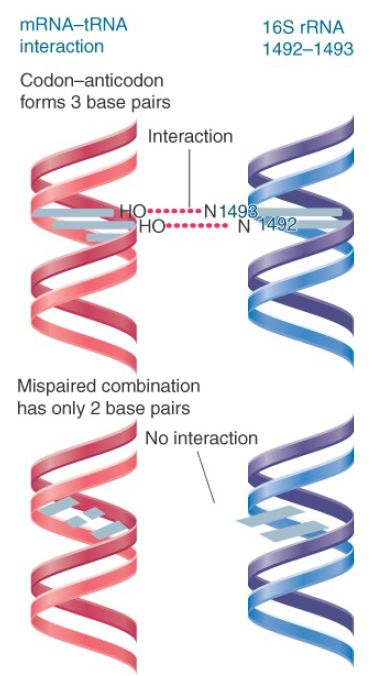
FIGURE 3. Codon–anticodon pairing supports interaction with adenines 1492 and 1493 of 16S rRNA, but mispaired tRNA–mRNA cannot interact.
A variety of bases in different positions of 16S rRNA are protected by tRNA in the P site; most likely the bases lie near one another in the tertiary structure. In fact, there are more contacts with tRNA when it is in the P site than when it is in the A site. This may be responsible for the increased stability of peptidyl-tRNA compared with aminoacyl-tRNA. This makes sense; once the tRNA has reached the P site, the ribosome has determined that it is correctly bound, whereas in the A site the assessment of binding is still being made. The 1400 region can be directly crosslinked to peptidyltRNA, which suggests that this region is a structural component of the P site.
The general conclusion of these results is that rRNA has many interactions with both tRNA and mRNA and that these interactions recur in each cycle of peptide bond formation.



|
|
|
|
دخلت غرفة فنسيت ماذا تريد من داخلها.. خبير يفسر الحالة
|
|
|
|
|
|
|
ثورة طبية.. ابتكار أصغر جهاز لتنظيم ضربات القلب في العالم
|
|
|
|
|
|
|
أصواتٌ قرآنية واعدة .. أكثر من 80 برعماً يشارك في المحفل القرآني الرمضاني بالصحن الحيدري الشريف
|
|
|