


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 17-5-2021
Date: 22-12-2015
Date: 25-3-2021
|
Initiation Involves Base Pairing Between mRNA and rRNA
KEY CONCEPTS
- An initiation site on bacterial mRNA consists of the AUG initiation codon preceded by the Shine–Dalgarno polypurine hexamer approximately 10 bases upstream.
- The rRNA of the 30S bacterial ribosomal subunit has a complementary sequence that base pairs with the Shine–Dalgarno sequence during initiation.
The signal for initiating a polypeptide chain is a special initiation codon that marks the start of the reading frame. Usually the initiation codon is the triplet AUG, but in bacteria GUG or UUG may also be used.
An mRNA may contain many AUG triplets, so how is the correct initiation codon recognized as the starting point for translation? The sites on mRNA where translation is initiated can be identified by binding the ribosome to mRNA under conditions that block elongation so that the ribosome remains at the initiation site. When ribonuclease is added to the blocked initiation complex, all the regions of mRNA outside the ribosome are degraded, but those actually bound to it are protected, as illustrated in Figure 1. The protected fragments can then be recovered and characterized.
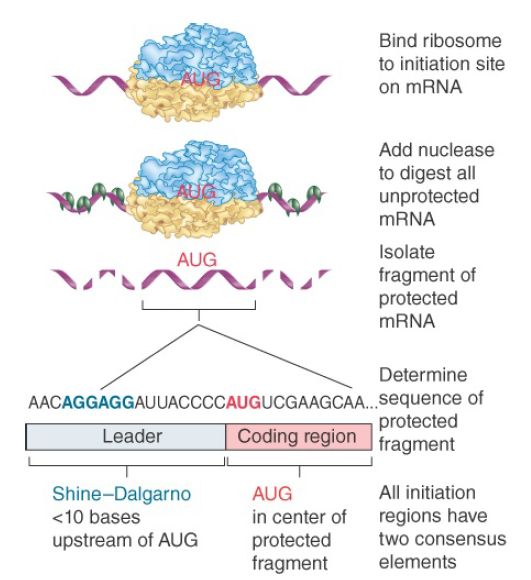
FIGURE 1. Ribosome-binding sites on mRNA can be identified by studying initiation complexes. They include the upstream Shine–Dalgarno sequence and the initiation codon.
The initiation sequences protected by prokaryotic ribosomes are approximately 30 bases long. The ribosome-binding sites of different bacterial mRNAs display two common features:
- The AUG (or less often, GUG or UUG) initiation codon is always included within the protected sequence.
- Approximately 10 bases upstream of the initiation codon is a sequence that corresponds to part or all of the hexamer:
5′ … A G G A G G … 3′
This polypurine stretch is known as the Shine–Dalgarno sequence. It is complementary to a highly conserved sequence close to the 3′ end of the 16S rRNA. (The extent of complementarity differs among individual mRNAs and ranges from a four-base core sequence GAGG to a nine-base sequence extending beyond each end of the hexamer.) Written in reverse
direction, the rRNA sequence is the hexamer:
3′ … U C C U C C … 5′
Does the Shine–Dalgarno sequence pair with its rRNA complement during mRNA–ribosome binding? Mutations of either sequence demonstrate its importance in initiation. Point mutations in the Shine–Dalgarno sequence can prevent an mRNA from being translated. In addition, the introduction of mutations into the complementary sequence in the rRNA is deleterious to the cell and changes the pattern of translation. The decisive confirmation of the base-pairing reaction is that a mutation in the Shine–Dalgarno sequence of an mRNA can be suppressed by a mutation in the rRNA that restores base pairing.
The sequence at the 3′ end of the rRNA is conserved among prokaryotes and eukaryotes, except that in all eukaryotes there is a deletion of the five-base sequence CCUCC that is the principal complement to the Shine–Dalgarno sequence. Base pairing does not appear to occur between eukaryotic mRNAs and the 18S rRNA. This is a significant difference between prokaryotes and eukaryotes in the mechanism of initiation.
In bacteria, a 30S subunit binds directly to a ribosome-binding site. As a result, the initiation complex forms at a sequence surrounding the AUG initiation codon. When the mRNA is polycistronic , each coding region starts with a ribosomebinding site.
The nature of bacterial gene expression means that translation of a polycistronic bacterial mRNA proceeds sequentially through each of its cistrons (coding regions). At the time when ribosomes attach to the first coding region, the subsequent coding regions have not yet been transcribed. By the time the second ribosomal binding site is available, translation through the first cistron is well under way.
What happens between the coding regions varies among individual polycistronic mRNAs. In most cases, the ribosomes probably bind independently at the beginning of each cistron. The most common series of events is illustrated in Figure 2. When synthesis of the first polypeptide terminates, the ribosomes leave the mRNA and dissociate into subunits. Then a new ribosome must assemble at the next coding region and begin translation of the next cistron.
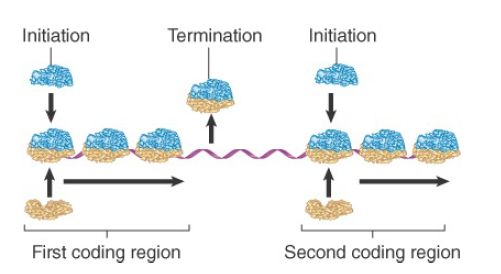
FIGURE 2. Initiation occurs independently at each cistron in a polycistronic mRNA. When the intercistronic region is longer than the span of sequence interacting with the ribosome, dissociation at the termination site is followed by independent reinitiation at the next cistron.
In some polycistronic bacterial mRNAs, translation between adjacent cistrons is directly linked, because ribosomes gain access to the initiation codon of the second cistron as they complete translation of the first cistron. This requires the distance between the two coding regions to be small. It may depend on the high local density of ribosomes, or the juxtaposition of termination and initiation sites could allow some of the usual intercistronic events to be bypassed. A ribosome physically spans about 30 bases of mRNA, so it can simultaneously contact a termination codon and the next initiation site if they are separated by only a few bases.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
المجمع العلمي ينظّم ندوة حوارية حول مفهوم العولمة الرقمية في بابل
|
|
|