


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 1-11-2015
Date: 30-10-2015
Date: 16-10-2015
|
Eukaryotic Chromosome
The deoxyribonucleic acid (DNA) of eukaryotic cells carries the blueprint for the biosynthesis of cellular proteins and the control of cellular assembly and regulation. If the entire DNA in a single human cell were stretched out straight and the strands representing all the chromosomes laid end-to-end, they would extend for well over 1 meter (3 feet). This meter of DNA must fit into a nucleus whose diameter is on the order of 10 microns (10-5 meter)! The dual problem of how to store this large amount of genetic information but also to keep it accessible for use and for faithful maintenance, copying, and distribution to daughter cells during cell division is solved by using proteins to package the DNA into chromosomes.
During the cell cycle, the cell grows (during G1 phase), replicates its DNA (during S phase), prepares for cell division (during G2 phase), and divides by mitosis (during M phase). During M phase, each chromosome is duplicated, and each replica remains attached to its original at the centromere portion of the chromosome. The two identical strands, called chromatids, wind up and become visible under the microscope at the beginning of mitosis. During the portion of mitosis known as metaphase, spindle fibers (which attach to the centromeres) jostle the chromatid pairs to the middle of the cell. The two chromatids are then pulled apart and segregated into different daughter cells, ensuring that each new cell has identical genetic information. The cells then enter G1 phase again. The combination of G1, S, and G2 is known as interphase. During interphase, the genes carried on the chromosomes are transcribed, to form proteins needed by the cell.
Various proteins act to stabilize DNA in interphase, while additional proteins are required to condense the chromosomes over a thousand fold to form the compact chromosomes required for mitosis and cell division. The sections that follow summarize key concepts concerning the structure of eukaryotic chromosomes.
Histones and Nucleosomes
Nearly all of the DNA in eukaryotic cells is complexed with a set of small basic proteins called histones. (As its name suggests, DNA is acidic, and is attracted to the basic histones.) The complexes form a repeating unit, the nucleosome, which consists of an octomeric disc of histones with about two turns of DNA wrapped around the outside. Thus, chromosomal DNA is organized as a string of nucleosome beads with a small amount of DNA connecting each bead. This first level of organization helps to compact the DNA so it can fit into the nucleus while still affording the necessary flexibility to fold the chromosome further; for example, in the condensation of chromosomes at metaphase.
The structure of the nucleosome is known at the atomic level through X-ray crystallography. The histone proteins interact extensively with one another to form the compact central disc of the nucleosomes, while specific amino acids have been identified that hold the DNA tightly onto the nucleosome surface. However, about fifteen to twenty-five amino acids at the end of each histone extend outside the compact limits of the central protein core. These tails are invisible in the X-ray structure of the nucleosome, indicating that they are relatively unstructured. (X-ray crystallography can only utilize structures with a high degree of order.) This indicates they can accommodate dynamic interactions with DNA or with adjacent nucleosomes in living chromosomes.

Immunofluorescence photomicrograph of the mitosis metaphase in an animal cell.
The sequence information encoded in DNA must be accessible to ribonucleic (RNA) polymerases in order to be useful as a template for transcription. Since the binding of DNA by histones interferes with this access, cells have evolved specific mechanism to destabilize nucleosomes in chromosome regions that must be transcribed. While the details of this important process are still being deciphered, it is clear that there are enzymes in eukaryotic nuclei that can modify nucleosome structure or the structure of individual histones to loosen the histone-DNA contacts, thereby making the DNA available for transcription.
One class of enzyme believed to modify nucleosomes for transcription is the histone acetyltransferases, which catalyze acetylation of specific lysines in the N-terminal tails of histones. Acetylation of lysines reduces the overall positive charge of the histone protein; since DNA has a net negative charge, histone acetylation may reduce the electrostatic forces holding the DNA on the nucleosome. This is thought to make the DNA more accessible to other DNA-binding proteins such as RNA polymerase.
In addition, some transcription regulatory proteins bind more easily to their DNA target sites if the nucleosomes associated with those sites are acetylated. A critical part of the transcription activation mechanism in eukaryotic cells appears to be the specific recruitment of nucleosome remodeling enzymes, such as histone acetyltransferases to promoters, thus allowing those promoters to be used more efficiently by RNA polymerase. Histone acetylation, therefore, can increase the transcription rate for a gene. Conversely, cells also possess histone deactylases. Histone deacetylases may be specifically recruited to shut off genes when they are no longer required.
Another type of histone modification is addition of a phosphate group called phosphorylation. Phosphorylation typically causes significant changes in protein structure and activity. Increased histone phosphorylation is correlated with chromosome condensation at the onset of mitosis. The mechanism by which phosphorylation promotes condensation is unclear, but may involve nucleosome-nucleosome interactions, or the binding of nonhistone proteins to nucleosomal DNA as part of the folding of chromosomes for metaphase.
30 Nanometer Fiber
The nucleosomal organization of DNA in chromosomes cannot fully account for the degree of compaction necessary to fit the genome into the compact nucleus. The nature of these additional levels of DNA folding is controversial, but is believed to include the coiling of nucleosome arrays to form a solenoidal structure. Such solenoids have been visualized in electron micrographs of eukaryotic chromosomes as fibers of 30 nanometers (a nanometer equals one billionth of a meter) in diameter, in contrast to the 10-nanometer diameter of the nucleosome particle itself. In somatic nuclei, the 30-nanometer fiber appears to be stabilized by a specific histone, his- tone H1, which interacts with the DNA-linking adjacent nucleosomes.
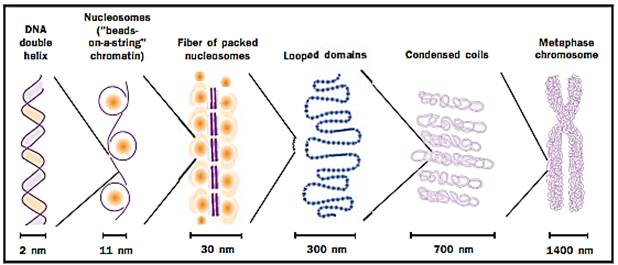
DNA is highly organized and has a structure that allows tight compaction while still allowing access for gene expression.
Domains and Higher Order Structures
Early electron micrograph images of eukaryotic metaphase chromosomes gave the impression of looped fibers extending out from the central axis of each chromatid. Subsequent analysis by microscopic and biochemical techniques suggests that stretches of chromosome approximately forty thousand to eighty thousand nucleotide pairs long may be anchored to a nuclear scaffold or matrix. These points of anchorage may serve to organize or spatially restrict chromosomes during interphase. These same anchor points may coalesce at metaphase to condense chromosomes for mitotic segregation.
Chromosomes exist to hold genes, of course, and some structural features of the chromosome may serve to separate genes from one another to help regulate transcription. Gene transcription in higher eukaryotes is controlled by regulatory elements that, in some cases, are located hundreds of thousands of nucleotides away from their target promoters. How can such elements be prevented from activating other nearby promoters? Experiments suggest that there are DNA sequences that act as boundaries or barriers to prevent the distant regulatory elements from one gene from contacting the promoters of genes located elsewhere on the same chromosome. In some cases, these genetic domain borders may be equivalent to the nuclear scaffold/matrix anchorage points, but in other cases these activities appear separable.
Telomeres, Telomerase, and Cancer
In his studies of chromosome structure, geneticist Herman Muller recognized that the natural ends of chromosomes were peculiar in that they could not be placed at internal sites in chromosomes, and that if they were detached (by breakage with ionizing radiation), the resulting chromosome behaved abnormally. He recognized the special properties of chromosome ends by giving them a special name: “telomeres.” Scientists now know that the ends of chromosomes have a unique structure and are maintained by a unique mechanism.

A karyotype of human male chromosomes (XY karyotype) with G banding.
The chromosomes of eukaryotic cells are linear DNA molecules. Because of this fact, and because of the mechanics of normal DNA replication by DNA-dependent DNA polymerases, a small amount of DNA at each end of every chromosome fails to be replicated with every cell cycle in somatic cells. If this loss occurred in the germ line as well, all eukaryotes would become extinct after a few generations, as important genes located near the chromosome ends would eventually be lost by the gradual chipping away at the ends. The major way that living cells offset this loss is by adding extra DNA onto one strand using a special enzyme for this purpose called “telomerase.” Telomerase is a ribonucleoprotein complex, consisting of an RNA- dependent DNA polymerase (also known as a reverse transcriptase) and an RNA molecule that serves as a template for DNA synthesis, giving rise to the characteristic repeated DNA sequence of most eukaryotic telomeres. (The fruit fly Drosophila is a notable exception to this; it uses transposable elements to maintain its telomeres.)
Telomerase activity in metazoans is found primarily in germ cells and at low levels in a few somatic tissues (stem cells that give rise to blood and skin cells that have to be replenished constantly throughout adult life). Normal animal somatic cells that are cultured in vitro usually lack telomerase activity. Such cells typically can divide only a finite number of times before they stop proliferating, go into a quiescent state, and eventually die, a process called senescence. Sencescence in cultured cells is correlated with loss of telomeric repeats. In general, cancer cells escape senescence and often can proliferate indefinitely in culture; this phenomenon, called immortalization, is accompanied by the activation of telomerase activity. Although cancer cells are often found to have unusually short telomeres, the length of their telomeres remains stable as the cells continue to proliferate. It is believed that telomerase activation in cancer is essential to continuous tumor growth and metastasis. Since most somatic cells have low or undetectable telomerase activity, drugs that specifically inactivate telomerase activity should be potent anticancer drugs with minimal side effects on healthy normal tissue.
Condensation and Decondensation
While chromosomes undergo cycles of condensation and decondensation with entry into and exit from mitosis during the cell cycle, some regions of chromosomes remain condensed throughout most of interphase. This chronically condensed material in the nuclei of all eukaryotic cells was recognized by German cytogeneticist Emil Heitz, who named it “heterochromatin” (in contrast with the “euchromatin,” or “true chromatin”), which disperses with the onset of interphase. The regions surrounding most eukaryotic centromeres is composed of heterochromatin.
Heterochromatin is distinguished from euchromatin by other properties. It replicates late in S phase while euchromatin replicates early in S, and it has the ability to silence euchromatic genes. Biochemical analysis shows that the DNA in heterochromatin is less accessible to a variety of DNA- binding proteins, suggesting that heterochromatin condensation inactivates regions of chromosomes by interfering with the accessibility of DNA for transcription. In mammalian females, one X chromosome is inactivated by heterochromatinization. This is thought to ensure that both males (who have only one X) and females (who have two) have equal “doses” of the many genes carried on the X chromosome.
Classes of DNA
The chromosomes of higher eukaryotes contain classes of DNA sequences that differ in the number of times they are presented in the genome. Much of the DNA in higher eukaryotes is unique, in the sense that the exact linear sequence of nucleotides is found only once per haploid chromosome complement. But some DNA sequences are found in a few dozen or a few hundred identical or nearly identical copies in each haploid chromosome set. These are considered “moderately repetitive” DNA sequences, and in most higher eukaryotes include the genes encoding the histones and the ribosomal RNA (rDNA), as well as certain classes of transposable elements. In the case of the repeated histone and rDNA, having many copies of these genes may be important at certain stages of development to allow biosynthesis of large amounts of histone proteins (during S phase) and ribosomal RNA (during ribosomal synthesis) in a short period of time.
The third broad class of DNA found in higher eukaryotic chromosomes is represented in many thousands of copies, and is thus termed “highly repetitive.” Because of the relative abundance and sequence homogeneity of highly repetitive DNA sequences, they were initially isolated from fragmented eukaryotic DNA as “sattelites” easily separated from the main mass of DNA. This satellite DNA includes tandem arrays—many copies, one right after another—of a 171-nucleotide pair repeat called “alphoid satellite.” Alphoid satellite DNA is found in tandem arrays of thousands of copies in the centromeres of all human chromosomes. The alphoid repeats are sufficient to confer centromeric properties on artificial human chromosomes. (The centromere region forms the “pinched waist” so characteristic of metaphase chromosomes, and is the site to which the spindle fibers attach to separate daughter chromatids in mitosis.
The function of other types of highly repetitive sequence DNA is unknown; indeed, some repetitive DNA sequences are thought to be “junk DNA,” present in chromosomes simply because there is no evolutionarily efficient way to eliminate it. Approximately 500,000 copies of a 300- nucleotide-pair sequence called an “Alu sequence” are found in the human genome. Unlike the alphoid satellite, Alu sequences are interspersed throughout all human chromosomes. Alu sequences are homologous to portions of the 7SL RNA, a structural component of the signal recognition particle that targets ribosomes to the endoplasmic reticulum. Alu sequences are probably relics of reverse transcription of this RNA into 7SL DNA, which then recombined randomly into chromosomes. Such dispersed repeated DNA sequences are potential sites for homologous recombination, not only between noncorresponding positions on the same chromosome or on different chromosomes. Indeed, recombination between Alu elements is probably responsible for some deletion or rearrangement of mutations leading to inherited human diseases, since Alu sequences are often found at deletion/rearrangement breakpoints.
Throughout all chromosomes of all living organisms, short, simple sequence repeats may be found. For example, short stretches of guanosine- cytosine base pairs, alternating adenosine-thymidine and cytosine-guanosine, occur randomly, both within and outside of protein-coding sequences, and are sometimes referred to as “microsatellite repeats.” In such regions, there is a higher tendency for the DNA polymerase to make errors by skipping a nucleotide or adding a couple of nucleotides. Such errors create sites of mismatched bases, which could lead to mutation—and cancer—if they are inherited by daughter cells after cell division. Most living cells have a way of detecting and correcting such mismatches shortly after they occur, using a mechanism termed “mismatch repair.” Patients that lack one of the components of the mismatch repair machinery have a much higher chance of being victims of certain types of cancers.
Identifying Chromosomes
Numbers and sizes of chromosomes vary widely in eukaryotes, and neither correlates with genome size. The classification of chromosomes within a given species was made possible initially by the used of stains that revealed variation in the DNA sequence composition along the length of the chromosome, resulting in a banded staining pattern characteristic for each chromosome. Using the criteria of overall chromosome length, relative centromere position and banding pattern, chromosomes of any species can be identified as a characteristic ordered set called a karyotype. With advent of molecular hybridization and extensive molecular cloning of unique- sequence DNAs, DNA sets representing sequences unique to individual chromosomes have been identified. By coupling the cloned DNA to fluorescent dyes and hybridizing the fluorescently labeled DNA directly to chromosomal preparations or whole cells, fluorescent in situ hybridization (FISH) enables rapid, efficient, and reliable identification of whole chromosomes or chromosome fragments. FISH has found widespread clinical application in the identification of chromosome rearrangements underlying inherited disease and many tumors.
Cytosine Methylation and Gene Regulation
When cellular DNA is first replicated, it consists of four nucleotide subunits: deoxyadenosine, deoxycytidine, deoxyguanosine, and thymidine. Following DNA replication, though, chemical modifications can occur to DNA. One of the most commonly encountered modifications found in the DNA of mammalian cells is the methylation of cytidine at carbon number 5 of the cytosine base. In human cells, about 3 to 5 percent of the cytosines are so methylated. The distribution of methylated sites is not uniform, but occurs only at cytosine residues that precede a guanosine (so-called CpG motifs, where the “p” symbolizes the intervening phosphate in the sugar-phosphate DNA backbone). Clusters of CpG dinucleotides—called CpG islands—preferentially occur near the promoters of many mammalian genes. When the cytosines in such islands are extensively methylated, the gene associated with that island is usually found to be transcriptionally silent. Thus, cytosine methylation is inversely correlated with gene expression. The mechanism of methylation-dependent silencing involves proteins that specifically recognize and bind to methylated DNA
Cytosine methylation is also found in plants, where it is also inversely correlated with gene activity. Interestingly, many fungi and insects have no detectable DNA methylation at all, yet they seem to be able to regulate their genes adequately. One theory is that DNA methylation arose in evolution as a secondary mechanism to ensure faithful gene silencing in organisms that undergo many cell divisions in development between fertilization and adulthood. It may also have evolved to inactivate certain types of viruses.
REFERNCES
Greider, Carol W., and Elizabeth H. Blackburn. “Telomeres, Telomerase and Cancer.” Scientific American 274, no. 2 (1996): 92-97.
Grunstein, Michael. “Histones as Regulators of Genes.” Scientific American 267, no. 4 (1992): 68-74B.
Moxon, E. Richard, and Christopher Wills. “DNA Microsatellites: Agents of Evolution?” Scientific American 280, no.1 (1999): 94-99.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
اتحاد كليات الطب الملكية البريطانية يشيد بالمستوى العلمي لطلبة جامعة العميد وبيئتها التعليمية
|
|
|