


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 6-12-2015
Date: 15-5-2016
Date: 31-12-2015
|
Noncoding RNAs Can Be Used to Regulate Gene Expression
KEY CONCEPTS
- Vast tracts of the eukaryotic genome are transcribed on both strands.
- A regulator RNA can function by forming a duplex region with a target RNA that may block initiation of translation, cause termination of transcription, or create a target for an endonuclease.
- Transcriptional interference occurs when an overlapping transcript on the same or opposite strand prevents transcription of another gene.
- Long ncRNAs (lncRNAs) are defined as longer than 200 nucleotides, without an open reading frame, and produced by RNA Pol II.
- Some noncoding RNAs (such as CUTs and PROMPTs) are often polyadenylated and very unstable.
- Noncoding RNAs can control the structure of the eukaryotic nucleus.
Noncoding RNAs (ncRNAs) and their genes, such as rRNA and tRNA, have been known since the 1950s. Whole families of new ncRNAs and their genes have been identified since then. These include snRNAs involved in splicing, snoRNAs involved in processing large RNAs such as rRNAs (see the chapter titled RNA Splicing and Processing), and microRNAs . These RNAs can generally be divided by size into large (rRNA size), medium (tRNA size), and microRNA sizes. This section focuses on the large-size class of ncRNAs, also called lncRNAs.
Experiments using both whole-genome tiling arrays (probing not just genes but whole genomes) and massive, whole-cell RNAsequencing experiments have shown that the vast majority of the eukaryotic genome is transcribed. This includes gene regions, of course, but surprisingly it also includes both the coding and noncoding strands of the genes, the regions between genes, telomeres, and centromeres. The estimate is that as much as 70% of human genes produce an antisense RNA. This pattern varies with the cell type and is presumably regulated. Transcription from the both the coding (sense) and noncoding (antisense) strands can result in noncoding RNAs with regulatory functions. Another ncRNA class is long intergenic noncoding RNA (lincRNA), as the name implies originating from intergenic regions, previously assumed to house no information. In addition to genes and antisense gene regions being transcribed, and the regions between genes being transcribed, promoters and enhancers are transcribed as well, giving rise to pRNAs (promoter RNA, sometimes called PROMPTs) and eRNAs (enhancer RNA).
A systematic, focused effort began a few years ago to examine the human genome in depth to understand its functional genomic content—called the Encyclopedia of DNA Elements (ENCODE) project. Shortly thereafter, the model organism ENCODE (modENCODE) projects were begun, focusing on the Caenorhabditis elegans and Drosophila melanogaster genomes.
The first phase of these projects has examined about 1% of the human genome and the entire C. elegans and Drosophila genomes.
At the start of the modENCODE project, C. elegans was known to have about 1000 ncRNAs. Data now support a model showing more than 21,000 ncRNAs called the 21k set. (Note that C.
elegans has about 19,000 classical genes, but what is the definition of a gene?) A second set, comprising about 7000 ncRNAs (called the 7k set) has been culled from the first by finetuning
the identification model. This in itself demonstrates the difficulty of distinguishing potentially genuine functional transcripts from accidental transcription events.
Base pairing offers a powerful means for one RNA to control the activity of another. Many cases have been identified in both prokaryotes and eukaryotes where a (usually rather short) singlestranded RNA base pairs with a complementary region of an mRNA, and as a result it prevents expression of the mRNA. One of the early illustrations of this effect was provided by an artificial situation in which antisense genes were introduced into eukaryotic cells.
Antisense genes are constructed by reversing the orientation of a gene with regard to its promoter, so that the “antisense” strand is transcribed into an antisense noncoding RNA, as illustrated in FIGURE 1. Synthesis of antisense RNA can inactivate a target RNA in either prokaryotic or eukaryotic cells. An antisense RNA is in effect an RNA regulator. Quantitation of the effect is not entirely reliable, but it seems that an excess (perhaps a considerable excess) of the antisense RNA may be necessary.
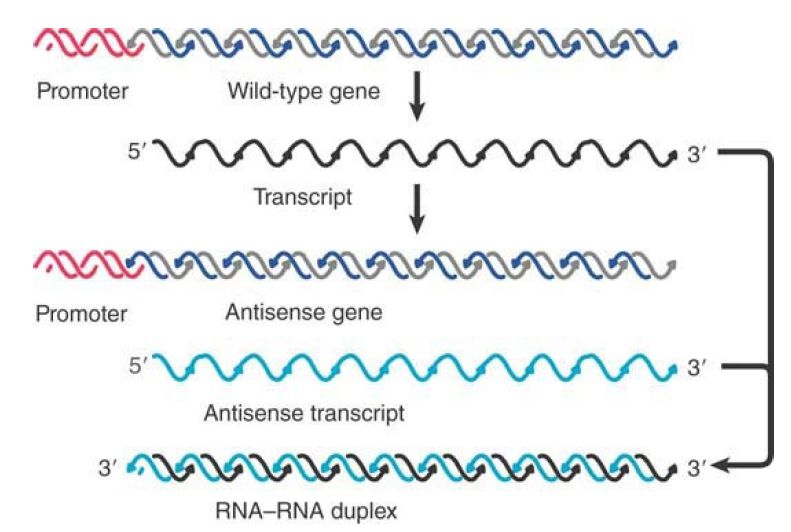
FIGURE 1.Antisense RNA can be generated by reversing the orientation of a gene with respect to its promoter and can anneal with the wild-type transcript to form duplex DNA.
At what stage does the antisense RNA inhibit expression? It could in principle prevent transcription of the authentic gene, processing of its RNA product, or translation of the messenger. Results with different systems show that the inhibition depends on formation of RNA–RNA duplex molecules, but this can occur either in the nucleus or in the cytoplasm. In the case of an antisense gene stably carried by a cultured cell, sense–antisense RNA duplexes form in the nucleus, preventing normal processing and/or transport of the sense RNA. In another case, injection of antisense RNA into the cytoplasm inhibits translation by forming duplex RNA in the 5′ region of the mRNA.
This technique offers a powerful approach for turning off genes at will; for example, the function of a regulatory gene can be investigated by introducing an antisense version. An extension of this technique is to place the antisense gene under the control of a promoter that is itself subject to regulation. The target gene can then be turned off and on by regulating the production of antisense RNA. This technique allows investigation of the importance of the timing of expression of the target gene.
Antisense RNA in eukaryotes has been known for some time. The first genome-sequencing projects demonstrated that nested genes (genes located within the introns of other genes) are widespread. They are more common than was first thought, comprising as much as 5% to 10% of genes. If the nested gene is transcribed from the opposite strand, then antisense RNA is produced. This head-tohead arrangement of a nested gene will also lead to transcriptional interference (TI), because both genes cannot be transcribed simultaneously.
Transcriptional interference has emerged as a significant mechanism of transcriptional regulation, and it can actually occur both when an interfering RNA is produced in an antisense orientation, as described earlier, or in the sense orientation. For example, the yeast SER3 gene (involved in serine biosynthesis) is normally repressed in the presence of serine and induced in its absence. It turns out that under serine-rich, repressive conditions, a noncoding RNA is expressed from the intergenic region upstream of the SER3 promoter and is transcribed from the same strand as SER3 across its promoter. This RNA (named for its gene, the SER3 regulatory gene, or SRG1) does not encode a protein, but its high expression ultimately serves to disrupt transcription initiation at the SER3 promoter. SRG1 is induced by serine; transcription by RNA pol II and the elongation factor Paf1 results in the recruitment of histone modification factors and the chromatin remodeling complex SWI/SNF, which then results in the deposition of a nucleosome on the SER3 promoter, preventing transcription. The end product of the biosynthetic pathway, serine, thus regulates SER3 by causing transcriptional interference at the SER3 promoter by a sense transcript. It is important to note that in transcriptional interference, it can be transcription per se, rather than the RNA product that is responsible for the regulatory effect.
A direct role for antisense RNA in transcription control has been demonstrated in the yeast Saccharomyces cerevisiae. The gene PHO84 is regulated in part by a class of noncoding RNAs called cryptic unstable transcripts, or CUTs. As shown in FIGURE 2, in addition to the promoter at the 5′ end of the gene, there is another promoter on the opposite strand that is unregulated. This promoter requires Set1 histone methyltransferase for activity and produces an antisense RNA. Under normal conditions, this RNA is rapidly degraded by the TRAMP (Transgenic Adenocarcinoma of the Mouse Prostate) complex and exosome RNase complexes as it is produced. In the absence of degradation or in aging cells, the antisense RNA persists. This antisense RNA, or CUT, works in trans to recruit histone deacetylase enzymes that remove acetate groups from histones, thereby causing the chromatin over the gene region to be
remodeled and condensed so that the gene can no longer be transcribed .
This is gene-specific remodeling directed by the antisense RNA and does not extend to the neighboring genes. The effect may also be brought about by a second exogenous copy of PHO84 on a plasmid in trans, called transcriptional gene silencing, or TGS, a phenomenon often seen in plants.
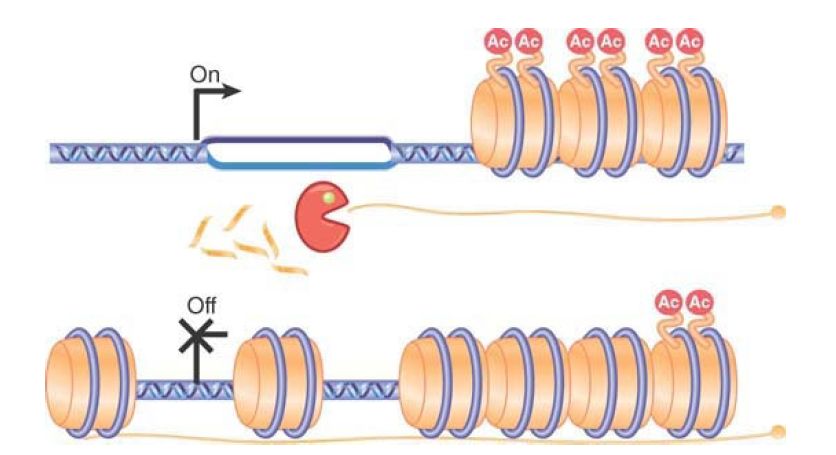
FIGURE 2 . PHO84 antisense RNA stabilization is paralleled by histone deacetylase recruitment, histone deacetylation, and PHO84 transcription repression. In wild-type cells, the RNA is rapidly degraded. In aging cells, antisense transcripts are stabilized and recruit the histone deacetylase to repress transcription. Data from J. Camblong, et al., Cell 131 (2007): 706–717.
Since this discovery, similar examples of ncRNAs that result in alteration of local chromatin structure have been described, such as a long RNA transcribed from the GAL1-10 locus that also results in histone deacetylation (as well as methylation) to promote GAL gene repression through chromatin remodeling. ncRNAs also prevent Ty retrotransposition through changes in chromatin structure in trans; this is reminiscent of the role of piRNAs in Drosophila (discussed in the chapter titled Regulatory RNA). This phenomenon may be quite widespread. In human HeLa cells, when a component of the RNA degradation machinery is disabled, vast amounts of upstream transcripts are observed from all three classes of active promoters (i.e., pRNAs, or PROMPTs). These RNAs are capped and polyadenylated at their 3′ end. Like CUTs in yeast, this RNA is very unstable. It can occur in both directions and may be related to the fact that open chromatin is available.
In addition to promoter-derived ncRNA (PROMPTs), enhancers are also transcribed and give rise to eRNAs. It has been proposed that these eRNAs (through base pairing with PROMPTs) can establish the necessary enhancer–promoter interactions necessary for initiating transcription.
Although some of these long ncRNAs are clearly derived from the promoters or gene body of classical genes, such as the PROMPTs and CUTs, others are derived from intergenic regions and are not associated with classical genes. One of the best examples, known for some time, is Xist . Ten different proteins bind to Xist RNA to exclude RNA Pol II and silence transcription. It also is responsible for recruiting the Polycomb repressor complex. (Interestingly, Xist itself is regulated by its antisense partner transcript, TsiX). Whereas Xist acts only in cis, on the X chromosome, others can act in trans, on multiple
chromosomes. In response to DNA damage, p53 acting as a transcription factor activates multiple lincRNAs. One of these, lincRNA-p21 , is itself targeted to multiple sites and acts as a transcription repressor.
Another lincRNA that is well characterized is the human HOTAIR, named because when discovered it was believed by many that this field of research was useless. It is transcribed from the developmental HOX C homeotic gene region but targets multiple genes on other chromosomes. At its target loci, it acts as a scaffold to assemble the Polycomb repressive complex 2 (PCR2) to reprogram chromatin structure and silence those genes that should be turned off.
HOTAIR expression has also been found to be deregulated in several cancers where it is associated with a poor prognosis. In general, ncRNAs can function in multiple ways, in cis, as with CUTs and PROMPTs, and in trans, as with HOTAIR. A second way to examine function is mechanistic. ncRNAs can work as antisense RNA, either by directly binding to its counterpart or by transcriptional interference. ncRNAs can function by binding and targeting a protein to a specific gene or region. Many ncRNAs work as scaffolds for chromatin modifiers and remodelers, either in cis or in trans. Alternatively, an ncRNA can bind a protein and act as an allosteric modifier.
It is becoming clear that lncRNAs play an important role beyond gene regulation. They also play a critical role in the overall structure of the nucleus itself, as shown in FIGURE 3. Chromosomes are not simply thrown into the nucleus randomly, but rather occupy specific nuclear domains called topologically associated domains (TADs). Homologous chromosomes have to be able to find each other at certain times in the meiotic cell cycle. This organization has been referred to as the chroperon.
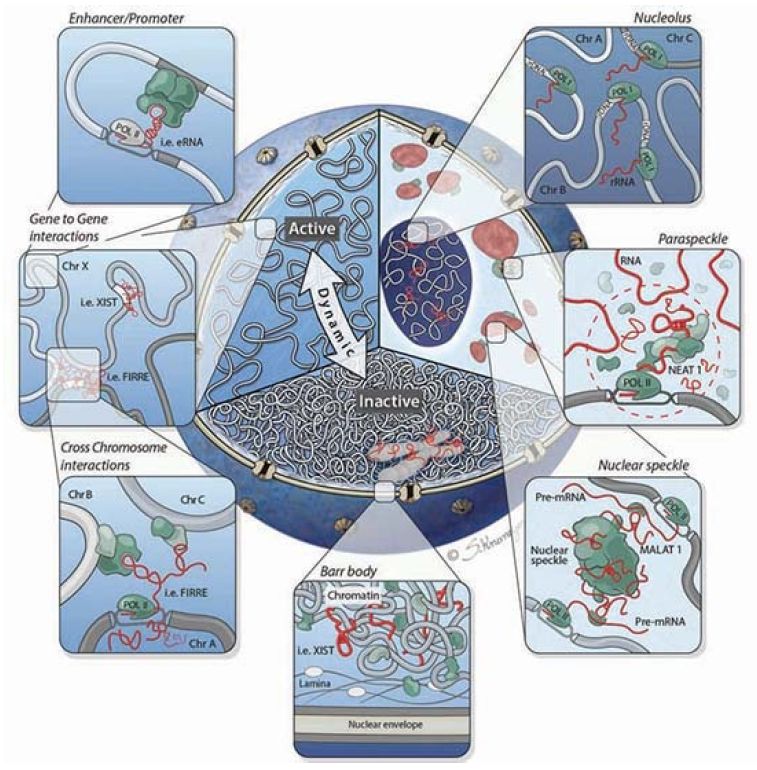
FIGURE 3. Cutaway of the nucleus highlighting three organizational levels: active (left) and inactive (bottom) regions, and nuclear bodies (right). Clockwise from right to left: The nucleolus is formed around actively transcribed rRNA sites; paraspeckles are formed by the Neat1 lncRNA; the Malat1 lncRNA is present within the nuclear speckle, and actively transcribed genes are repositioned close to nuclear speckles; the inactive X chromosome (Barr body) is coated by the Xist lncRNA and dynamically repositioned from the active to inactive compartments where it is localized to the periphery of the nucleus; lncRNAs can mediate gene-gene interactions across chromosomes (bottom panel inset) and within chromosomes (top panel inset).



|
|
|
|
4 أسباب تجعلك تضيف الزنجبيل إلى طعامك.. تعرف عليها
|
|
|
|
|
|
|
أكبر محطة للطاقة الكهرومائية في بريطانيا تستعد للانطلاق
|
|
|
|
|
|
|
أصواتٌ قرآنية واعدة .. أكثر من 80 برعماً يشارك في المحفل القرآني الرمضاني بالصحن الحيدري الشريف
|
|
|