


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 11-5-2016
Date: 7-6-2021
Date: 8-5-2016
|
Heterochromatin Propagates from a Nucleation Event
KEY CONCEPTS
- Heterochromatin is nucleated at a specific sequence, and the inactive structure propagates along the chromatin fiber.
- Heterochromatin nucleation is caused by proteins binding to specific sequences.
- Genes within regions of heterochromatin are inactivated.
- The length of the inactive region varies from cell to cell; as a result, inactivation of genes in this vicinity causes position-effect variegation.
- The two states of gene expression (on or off) affect phenotype based on the variable positions.
- Similar spreading effects occur at telomeres and at the silent cassettes in yeast mating-type loci.
An interphase nucleus contains both euchromatin and heterochromatin. The condensation state of heterochromatin is close to that of mitotic chromosomes. Heterochromatin is generally inert. It remains condensed in interphase, is transcriptionally repressed, replicates late in S phase, and may be localized to the nuclear periphery. Centromeric heterochromatin typically consists of repetitive satellite DNAs; however, the formation of heterochromatin is not rigorously defined by sequence. When a gene is transferred, either by a chromosomal translocation or by transfection and integration, into a position adjacent to heterochromatin, it may become inactive as the result of its new location, implying that it has become heterochromatic.
Such inactivation is the result of an epigenetic effect . It may differ between individual cells in an organism, in which case it results in the phenomenon of position-effect variegation (PEV), in which genetically identical cells have different phenotypes. Genes affected by PEV have two states—active or silenced—depending on their position relative to the boundary of heterochromatin, which can lead to variegated phenotypes. This has been well characterized in Drosophila. FIGURE 1 shows an example of PEV in the fly eye. Some of the regions in the eye lack color, whereas others are red. This is because the white gene (required to develop red pigment) is inactivated by adjacent heterochromatin in some cells but remains active in others.

FIGURE 1. Position-effect variegation (PEV) in eye color results when the white gene is integrated near heterochromatin. Cells in which white is inactive give patches of white, whereas cells in which white is active give red patches. The severity of the effect is determined by the closeness of the integrated gene to heterochromatin.
Photo courtesy of Steven Henikoff, Fred Hutchinson Cancer Research Center.
The explanation for this effect is shown in FIGURE 2. Inactivation spreads from heterochromatin into the adjacent region for a variable distance. In some cells it goes far enough to inactivate a nearby gene, whereas in others it does not. This happens at a certain point in embryonic development, and after that point the state of the gene is stably inherited by all the progeny cells. Cells descended from an ancestor in which the gene was inactivated form patches corresponding to the phenotype of loss of function (in the case of white, the absence of color).
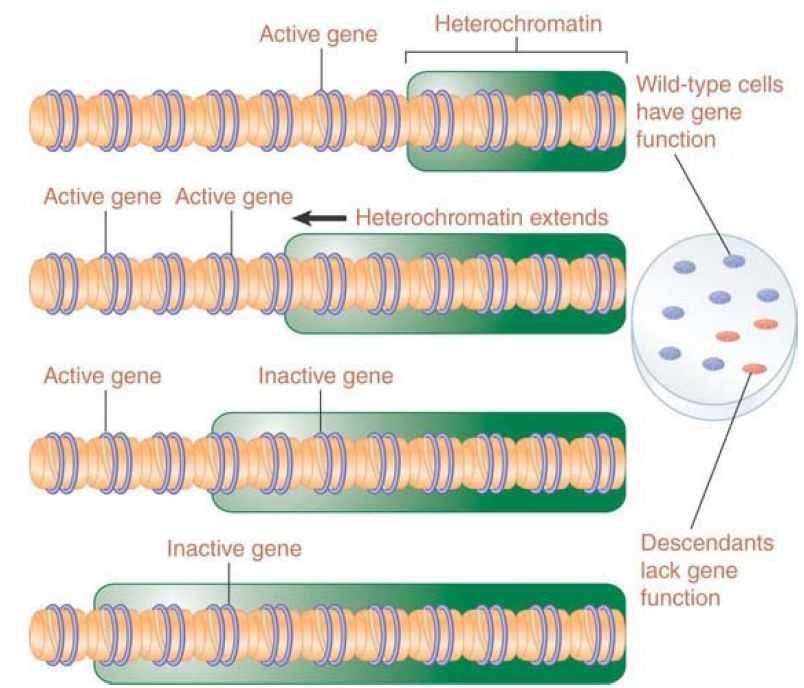
FIGURE 2. Extension of heterochromatin inactivates genes. The probability that a gene will be inactivated depends on its distance from the heterochromatin region.
The closer a gene lies to heterochromatin, the higher the probability that it will be inactivated. This is due to the fact that formation of heterochromatin is typically a two-stage process: A nucleation event occurs at a specific sequence or region (triggered by binding of a protein that recognizes the DNA sequence or other identifiers in the region), and then the inactive structure propagates along the chromatin fiber. The distance by which the inactive structure extends is not precisely determined and may be stochastic, being influenced by parameters such as the quantities of limiting protein components. Another factor that may affect the spreading process is the activation of promoters in the region; an active promoter may inhibit spreading. Genes near heterochromatin are more likely to be inactivated; however, insulators can protect a transcriptionally active region by preventing heterochromatin from spreading .
The effect of telomeric silencing in yeast is analogous to PEV in Drosophila; genes translocated to a telomeric location show the same sort of variable loss of activity. This results from a spreading effect that propagates from the telomeres. In this case, the binding of the Rap1 protein to telomeric repeats triggers the nucleation event, which results in the recruitment of heterochromatin proteins, as described in the next section, Heterochromatin Depends on Interactions with Histones.
In addition to the telomeres, heterochromatin is nucleated at two other sites in yeast. Yeast mating type is determined by the activity of a single active locus (MAT), but the genome contains two other copies of the mating-type sequences (HML and HMR), which are maintained in an inactive form. The silent loci HML and HMR nucleate heterochromatin via binding of several proteins (rather than the single protein, Rap1, required at telomeres), which then leads to propagation of heterochromatin, similar to that at telomeres. Heterochromatin in yeast exhibits features typical of heterochromatin in other species, such as transcriptional inactivity and self-perpetuating protein structures superimposed on nucleosomes (which are generally deacetylated). The only notable difference between yeast heterochromatin and that of most other species is that histone methylation in yeast is not associated with silencing, whereas specific sites of histone methylation are a key feature of heterochromatin formation in most eukaryotes.



|
|
|
|
تفوقت في الاختبار على الجميع.. فاكهة "خارقة" في عالم التغذية
|
|
|
|
|
|
|
أمين عام أوبك: النفط الخام والغاز الطبيعي "هبة من الله"
|
|
|
|
|
|
|
المجمع العلمي ينظّم ندوة حوارية حول مفهوم العولمة الرقمية في بابل
|
|
|