


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية| Newly Synthesized RNAs Are Checked for Defects via a Nuclear Surveillance System |
|
|
|
Read More
Date: 11-11-2020
Date: 21-12-2015
Date: 4-5-2016
|
Newly Synthesized RNAs Are Checked for Defects via a Nuclear Surveillance System
KEY CONCEPTS
- Aberrant nuclear RNAs are identified and destroyed by a surveillance system.
- The nuclear exosome functions both in the processing of normal substrate RNAs and in the destruction of aberrant RNAs.
- The yeast TRAMP complex recruits the exosome to aberrant RNAs and facilitates its 3′ to 5′ exonuclease activity.
- Substrates for TRAMP-exosome degradation include unspliced or aberrantly spliced pre-mRNAs and improperly terminated RNA Pol II transcripts lacking a poly(A) tail.
- The majority of RNA Pol II transcripts may be cryptic unstable transcripts (CUTs) that are rapidly destroyed in the nucleus.
All newly synthesized RNAs are subject to multiple processing steps after they are transcribed . At each step, errors may be made. Whereas DNA errors are repaired by a variety of repair systems (see the chapter titled Repair Systems), detectable errors in RNA are dealt with by destroying the defective RNA. RNA surveillance systems exist in both the nucleus and cytoplasm to handle different kinds of problems. Surveillance involves two kinds of activities: one to identify and tag the aberrant substrate RNA, and another to destroy it.
The destroyer is the nuclear exosome. The nuclear exosome core is almost identical to the cytoplasmic exosome, though it interacts with different protein cofactors. It removes nucleotides from targeted RNAs by 3′ to 5′ exonuclease activity. The nuclear exosome has multiple functions involving RNA processing of some noncoding RNA transcripts (snRNA, snoRNA, and rRNA) and complete degradation of aberrant transcripts. The exosome is recruited to its processing substrates by protein complexes that recognize specific RNA sequences or RNA–RNP structures. For example, Nrd1–Nab3 is a sequence-specific protein dimer that recruits the exosome to normal sn/snoRNA processing substrates.
This protein pair binds to GUA[A-G] and UCUU elements, respectively. The Nrd1–Nab3 cofactor is also involved in transcription termination of these nonpolyadenylated Pol II–transcribed RNAs, suggesting that the processing exosome may be recruited directly to the site of their synthesis.
Aberrantly processed, modified, or misfolded RNAs require other protein cofactors for identification and exosome recruitment. The major nuclear complex performing this function in yeast is called TRAMP (an acronym for the component proteins), and it exists in at least two forms, differing in the type of poly(A) polymerase present. The TRAMP complex acts in several ways to effect degradation:
- It interacts directly with the exosome, stimulating its exonuclease activity.
- It includes a helicase, which is probably required to unwind secondary structure and/or move RNA-binding proteins from structured RNP substrates during degradation.
- It adds a short 3′ oligo(A) tail to target substrates. The oligo(A) tail is thought to make the targeted RNP a better substrate for the degradation machinery in the same way that the oligo(A) tail functions in bacteria.
FIGURE 1. summarizes the roles of TRAMP and the exosome. It has become clear that RNA degradation in bacteria and archaea and nuclear RNA degradation in eukaryotes are evolutionarily related processes. Their similarity suggests that the ancestral role of polyadenylation was to facilitate RNA degradation, and that poly(A) was later adapted in eukaryotes for the oddly reverse function of stabilizing mRNAs in the cytoplasm.
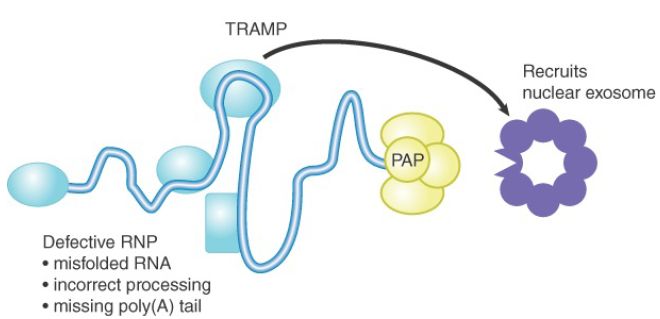
FIGURE 1.The role of TRAMP and the exosome in degrading aberrant nuclear RNAs. Defective RNPs are tagged by protein cofactors, which then recruit the nuclear exosome. The cofactor in yeast cells is the complex TRAMP. The poly(A) polymerase (PAP, or Trf4) in TRAMP adds a short poly(A) tail to the 3′ end of the targeted RNA.
What are the substrates for TRAMP–exosome degradation? The TRAMP complex is remarkable in that it recognizes a wide variety of aberrant RNAs synthesized by all three transcribing polymerases. It is not known how this is accomplished given that the targeted RNAs share no recognizably common features. Some researchers favor a kinetic competition model, hypothesizing that RNAs that do not get processed and assembled into final RNP form in a timely manner will become substrates for exosome degradation. This mechanism avoids the need to posit specific recognition of innumerable possible defects.
What kinds of abnormalities condemn pre-mRNAs to nuclear destruction? Two kinds of substrates have been identified. One type is unspliced or aberrantly spliced pre-mRNAs. Components of the spliceosome retain such transcripts either until they are degraded by the exosome or until proper splicing is completed, if possible. It is thought that the kinetic competition model probably applies here, too. A pre-mRNA that is not efficiently spliced and packaged is at increased risk of being accessed by the exosome degradation machinery. The basis for recognition of aberrantly spliced pre-mRNAs is not known. The second type of pre-mRNA substrate is one that has been improperly terminated, lacking a poly(A) tail. Whereas polyadenylation is protective in true mRNAs, it may actually be destabilizing for cryptic unstable transcripts (CUTs). These non-protein-coding RNAs (also discussed in the Regulatory RNA chapter) are transcribed by RNA Pol II and do not encode recognizable genes; however, they frequently overlap with (and sometimes regulate) protein-coding genes. These transcripts are polyadenylated by a component of the TRAMP complex (Trf4).
They are distinguished from other transcripts of unknown function by their extreme instability, normally being degraded by the TRAMP–exosome complex immediately after synthesis, possibly targeted by the Trf4-dependent polyadenylation. In fact, the existence of these transcripts was first convincingly demonstrated in yeast strains with impaired nuclear RNA degradation. More than three-quarters of RNA Pol II transcripts may be composed of noncoding RNAs and be subject to rapid degradation by the exosome! Some CUTs appear to arise from spurious transcription initiation, and the short-lived RNA products themselves typically do not appear to have a function (i.e., these RNAs do not typically act in trans). However, some examples indicate that the transcription process itself may play a role in regulating nearby or overlapping coding genes .



|
|
|
|
مقاومة الأنسولين.. أعراض خفية ومضاعفات خطيرة
|
|
|
|
|
|
|
أمل جديد في علاج ألزهايمر.. اكتشاف إنزيم جديد يساهم في التدهور المعرفي ؟
|
|
|
|
|
|
|
العتبة العباسية المقدسة تنظّم دورةً حول آليّات الذكاء الاصطناعي لملاكاتها
|
|
|