


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 30-11-2015
Date: 16-3-2021
Date: 5-6-2021
|
How Does RNA Polymerase Find Promoter Sequences
KEY CONCEPTS
- The rate at which RNA polymerase binds to promoters can be too fast to be accounted for by simple diffusion.
- RNA polymerase binds to random sites on DNA and exchanges them with other sequences until a promoter is found.
RNA polymerase must find promoters within the context of the genome. How are promoters distinguished from the 4 × 106 bp that comprise the rest of the E. coli genome? FIGURE 1. illustrates simple models for how RNA polymerase might find promoter sequences from among all the sequences it can access. RNA polymerase holoenzyme locates the chromosome by random diffusion and binds sequence nonspecifically to the negatively charged DNA. In this mode, holoenzyme dissociates very rapidly. Diffusion sets an upper limit for the rate constant for associating with a 75-bp target of less than 108 M-1 sec-1 . The actual forward rate constant for some promoters in vitro, however, appears to be approximately 108 M-1 sec-1 , at or above the diffusion limit. Making and breaking a series of complexes until (by chance) RNA polymerase encounters a promoter and progresses to an open complex capable of making RNA would be a relatively slow process. Thus, the time required for random cycles of successive association and dissociation at loose binding sites is too great to account for the way RNA polymerase finds its promoter. RNA polymerase must therefore use some other means to seek its binding sites.
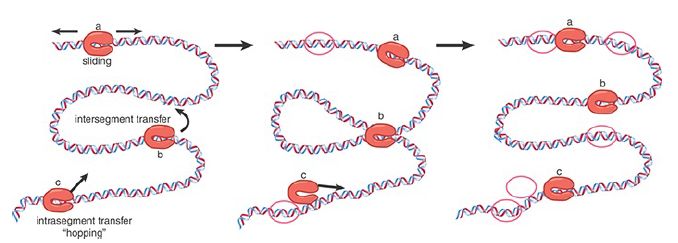
FIGURE 1 Proposed mechanisms for how RNA polymerase finds a promoter: (a) sliding, (b) intersegment transfer, (c) intradomain association and dissociation or hopping. Data from C. Bustamante, et al., J. Biol. Chem. 274 (1999): 16665–16668.
Figure 1 shows that the process is likely to be sped up because the initial target for RNA polymerase is the whole genome, not just a specific promoter sequence. By increasing the target size, the rate constant for diffusion to DNA is correspondingly increased and is no longer limiting. How does the enzyme move from a random binding site on DNA to a promoter? Considerable evidence suggests that at least three different processes contribute to the rate of promoter search by RNA polymerase. First, the enzyme may move in a one-dimensional random walk along the DNA (“sliding”). Second, given the intricately folded nature of the chromosome in the bacterial nucleoid, having bound to one sequence on the chromosome, the enzyme is now closer to other sites, reducing the time needed for dissociation and rebinding to another site (“intersegment transfer” or “hopping”). Third, while bound nonspecifically to one site, the enzyme may exchange DNA sites until a promoter is found (“direct transfer”).



|
|
|
|
علامات بسيطة في جسدك قد تنذر بمرض "قاتل"
|
|
|
|
|
|
|
أول صور ثلاثية الأبعاد للغدة الزعترية البشرية
|
|
|
|
|
|
|
مدرسة دار العلم.. صرح علميّ متميز في كربلاء لنشر علوم أهل البيت (عليهم السلام)
|
|
|