


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 10-12-2015
Date: 6-12-2015
Date: 24-3-2021
|
Yeast Ty Elements Resemble Retroviruses
KEY CONCEPTS
- Ty transposons have an organization similar to that of endogenous retroviruses.
- Ty transposons are retrotransposons (with a reverse transcriptase activity) that transpose via an RNA intermediate.
Ty elements comprise a family of dispersed repetitive DNA sequences that are found at different sites in different strains of yeast. Ty is an abbreviation for “transposon yeast.” Five types of Ty elements in yeast (Ty1–Ty5) have been identified. All are LTR retrotransposons, with characteristic LTRs and gag and pol genes with homology to those encoded by retroviruses. These elements are representative of two of the major classes of retrotransposons in eukaryotes, the Ty1/copia class (Ty1, Ty2, Ty4, and Ty5) and the Ty3/gypsy class. Each class is phylogenetically distinct, and each contains a characteristic order of open reading frames.
In the yeast Saccharomyces cerevisiae, Ty1 is the most abundant and the most well-characterized retroelement. A Ty1 transposition event creates a characteristic footprint: 5 bp of target DNA are repeated on either side of the inserted Ty1 element. Under most circumstances the frequency of Ty1 transposition is lower than that of most bacterial transposons, about 10–7 to 10–8 , but it can be increased by a variety of factors that stress the organism, such as mutagens and nutrient depletion.
The general organization of Ty1 elements is illustrated in FIGURE 1. Each element is 5.9 kb long; the last 334 bp at each end constitute LTRs, called delta (δ) for historical reasons but referred to here simply as LTRs. Individual Ty1 have many changes from the prototype of their class, including base-pair substitutions, insertions, and deletions. The typical yeast genome has about 30 copies of Ty1 and 13 copies of the closely related Ty2. In addition, there are around 180 independent solo Ty1/Ty2 LTRs.
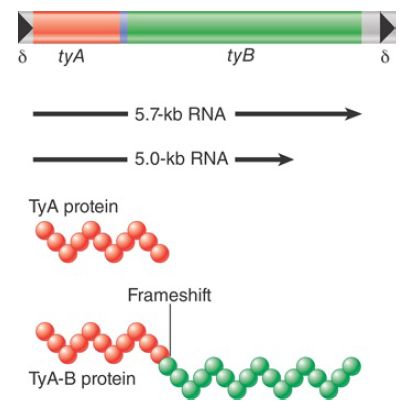
FIGURE 1. Ty elements terminate in short direct repeats and are transcribed into two overlapping RNAs. They have two reading frames, with sequences related to the retroviral gag and pol genes.
The LTR sequences also show considerable heterogeneity, although the two repeats of an individual Ty1 element are often identical or at least very closely related. The LTR sequences associated with Ty1 elements show greater conservation of sequence than the solo LTRs. This is because transposition of Ty1 elements, like replication of retroviruses, involves duplication of the LTRs (discussed in the following paragraphs). Thus, recently inserted elements carry identical LTRs, but solo LTRs diverge over time due to random mutations.
The Ty1 element is transcribed into two poly(A)+ RNA species, which constitute as much as 8% of the total mRNA of a haploid yeast cell. Both species initiate within a promoter in the LTR at the left end. One terminates after 5 kb; the other terminates after 5.7 kb, within the LTR sequence at the right end.
The sequence of the Ty1 element has two open reading frames. These frames are expressed in the same direction, but are read in different phases and overlap by 13 amino acids. TyA is related to retroviral gag genes and encodes a capsid protein. TyB contains regions that have homologies with reverse transcriptase, protease, and integrase sequences of retroviruses.
The organization and functions of TyA and TyB are analogous to the behavior of the retroviral gag and pol functions. The reading frames TyA and TyB are expressed in two forms. The TyA protein represents the TyA reading frame and terminates at its end. The TyB reading frame, however, is expressed only as part of a joint protein, in which the TyA region is fused to the TyB region by a specific frameshift event that allows the termination codon to be bypassed. (This is analogous to gag-pol translation in retroviruses.)
Recombination between Ty1 elements seems to occur in bursts; when one event is detected, the probability of finding others is increased. Gene conversion occurs between Ty1 elements at different locations, with the result that one element is “replaced” by the sequence of the other.
Ty elements can be deleted via homologous recombination between the directly repeated LTR sequences. The large number of solo LTR elements may be footprints of such events. A deletion of this nature may be associated with reversion of a mutation caused by the insertion of Ty; the level of reversion may depend on the exact LTR sequences left behind and the nature of the insertion site.
A paradox is that both LTRs have the same sequence, yet a promoter is active in the LTR at one end and a terminator is active in the LTR at the other end. (A similar feature is found in other
transposable elements, including the retroviruses.) Ty elements are classic retrotransposons in that they transpose through an RNA intermediate. An ingenious protocol used to detect this event is illustrated in FIGURE 2. An intron was inserted into an element to generate a unique Ty sequence. This sequence was placed under the control of a GAL promoter on a plasmid and introduced into yeast cells. Transposition results in the appearance of multiple copies of the transposon in the yeast genome, but the copies all lack the intron.
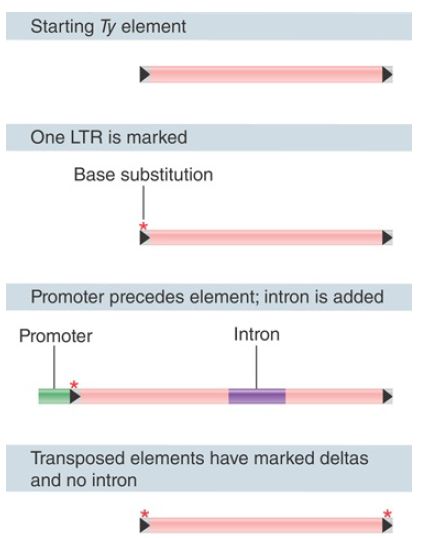
FIGURE 2. A unique Ty element, engineered to contain an intron, transposes to give copies that lack the intron. The copies possess identical terminal repeats, which are generated from one of the termini of the original Ty element.
We know of only one way to remove introns: RNA splicing. This suggests that transposition occurs by the same mechanism as with retroviruses. The Ty element is transcribed into an RNA that is recognized by the splicing apparatus. The spliced RNA is recognized by a reverse transcriptase and regenerates a duplex DNA copy, which is then integrated back into the genome using the integrase protein.
The analogy with retroviruses extends further. The original Ty1 element has a difference in sequence between its two LTRs. The transposed elements possess identical delta sequences, however, which are derived from the 5′ delta of the original element.
Transposition is controlled by genes within the Ty1 element. The GAL promoter used to control transcription of the marked Ty1 element is inducible: It is turned on by the addition of galactose. Induction of the promoter has two effects. It is necessary to activate transposition of the marked element, and its activation also increases the frequency of transposition of the other Ty1 elements on the yeast chromosome. This implies that the products of the Ty1 element can act in trans on other elements (actually on their RNAs).
The Ty element does not give rise to infectious particles; instead, virus-like particles (VLPs) with icosahedral features accumulate within the cells in which transposition has been induced. The particles contain full-length RNA, double-stranded DNA, reverse transcriptase activity, and a TyB product with integrase activity and are associated with RNA processing bodies (P bodies). The TyA product is cleaved like a gag precursor to produce the mature core proteins of the VLP.
Not all of the Ty1 elements in any yeast genome are active: Some have lost the ability to transpose (and are analogous to inert endogenous proviruses). These “dead” elements retain LTRs, though, and as a result they provide targets for transposition in response to the proteins synthesized by an active element.



|
|
|
|
5 علامات تحذيرية قد تدل على "مشكل خطير" في الكبد
|
|
|
|
|
|
|
تستخدم لأول مرة... مستشفى الإمام زين العابدين (ع) التابع للعتبة الحسينية يعتمد تقنيات حديثة في تثبيت الكسور المعقدة
|
|
|