


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 14-12-2015
Date: 15-11-2020
Date: 10-12-2015
|
Base Excision Repair Systems Require Glycosylases
KEY CONCEPTS
- Base excision repair is triggered by directly removing a damaged base from DNA.
- Base removal triggers the removal and replacement of a stretch of polynucleotides.
- The nature of the base removal reaction determines which of two pathways for base excision repair is activated.
- The polδ/ε pathway replaces a long polynucleotide stretch; the polβ pathway replaces a short stretch.
- Uracil and alkylated bases are recognized by glycosylases and removed directly from DNA.
- Glycosylases and photolyase act by flipping the base out of the double helix, where, depending on the reaction, it is either removed or modified and returned to the helix.
Base excision repair is similar to the nucleotide excision repair pathways described in the previous section. The process usually starts in a different way, however, with the removal of an individual damaged base. This serves as the trigger to activate the enzymes that excise and replace a stretch of DNA, including the damaged site.
Enzymes that remove bases from DNA are called glycosylases and lyases. FIGURE 1 shows that a glycosylase cleaves the bond between the damaged or mismatched base and the deoxyribose. FIGURE 2 shows that some glycosylases are also lyases that can take the reaction a stage further by using an amino (NH ) group to attack the deoxyribose ring. This is usually followed by a reaction that introduces a nick into the polynucleotide chain. FIGURE 3 shows that the exact form of the pathway depends on whether the damaged base is removed by a glycosylase or lyase.

FIGURE 1. A glycosylase removes a base from DNA by cleaving the bond to the deoxyribose.

FIGURE 2. A glycosylase hydrolyzes the bond between base and deoxyribose (using H2 O), but a lyase takes the reaction further by opening the sugar ring (using NH2 ).
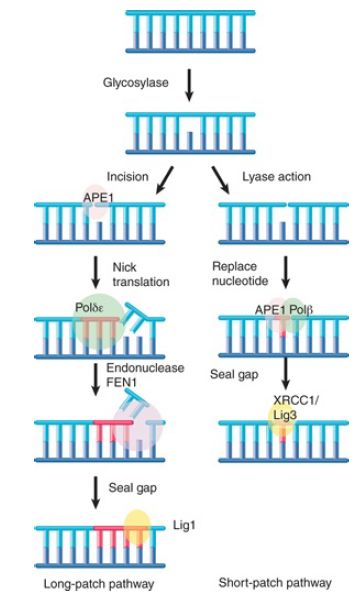
FIGURE 3. Base removal by glycosylase or lyase action triggers mammalian excision repair pathways.
Glycosylase action is followed by the endonuclease APE1, which cleaves the polynucleotide chain on the 5′ side. This, in turn, attracts a replication complex that includes DNA polymerase δ/ε and ancillary components. The replication complex performs a short synthesis reaction extending for 2 to 10 nucleotides. The displaced material is removed by the flap endonuclease (FEN1).
The enzyme ligase 1 seals the chain. This is called the long-patch pathway. (Note that these names refer to mammalian enzymes, but the descriptions are generally applicable for all eukaryotes.) When the initial removal involves lyase action, the endonuclease APE1 instead recruits DNA polymerase β to replace a single nucleotide. The nick is then sealed by the ligase XRCC1/ligase 3. This is called the short-patch pathway.
Several enzymes that remove or modify individual bases in DNA use a remarkable reaction in which a base is “flipped” out of the double helix. This type of interaction was first demonstrated for methyltransferases—enzymes that add a methyl group to cytosine in DNA. This base-flipping mechanism places the base directly into the active site of the enzyme, where it can be modified and returned to its normal position in the helix or, in the case of DNA damage, immediately excised. Alkylated bases (typically in which a methyl group has been added to a base) are removed by this mechanism. A human enzyme, alkyladenine DNA glycosylase (AAG), recognizes and removes a variety of alkylated substrates, including 3-methyladenine, 7-methylguanine, and hypoxanthine.
FIGURE 4. shows the structure of AAG bound to a methylated adenine, in which the adenine is flipped out and bound in the glycosylase’s active site.

FIGURE 4. Crystal structure of the DNA repair enzyme alkyladenine DNA glycosylase (AAG) bound to a damaged base (3-methyladenine). The base (black) is flipped out of the DNA double helix (blue) and into AAG’s active site (orange and green).
Courtesy of CDC.
By contrast with this mechanism, 1-methyl-adenine is corrected by an enzyme that uses an oxygenating mechanism (encoded in E. coli by the gene alkB, which has homologs in numerous eukaryotes, including three human genes). The methyl group is oxidized to a CH 2OH group, and then the release of the HCHO moiety (formaldehyde) restores the structure of adenine. A very interesting discovery is that the bacterial enzyme, and one of the human enzymes, can also repair the same damaged base in RNA. In the case of the human enzyme, the main target may be ribosomal RNA. This is the first known repair event with RNA as a target.
One of the most common reactions in which a base is directly removed from DNA is catalyzed by uracil-DNA glycosylase. Uracil typically only occurs in DNA because of spontaneous deamination of cytosine. It is recognized by the glycosylase and removed. The reaction is similar to that shown in Figure 4.: The uracil is flipped out of the helix and into the active site in the glycosylase. It appears that most or all glycosylases and lyases (in both prokaryotes and eukaryotes) work in a similar way.
Another enzyme that uses base flipping is the photolyase that reverses the bonds between pyrimidine dimers . The pyrimidine dimer is flipped into a cavity in the enzyme. Close to this cavity is an active site that contains an electron donor, which provides the electrons to break the bonds. Energy for the reaction is provided by light in the visible wavelength. Although most prokaryotic and eukaryotic species possess photolyase, placental mammals (but not marsupials) have lost this activity.
The common feature of these enzymes is the flipping of the target base into the enzyme structure. Recent work has shown that Rad4, the yeast XPC homolog (the protein that recognizes UV damage and other lesions during nucleotide excision repair), uses an interesting variation on this theme. Rad4 flips out the two adenine bases that are complementary to the linked thymines in a pyrimidine dimer, rather than flipping out the damaged pyrimidine dimer itself. In fact, it is believed that the ease with which these unpaired adenines are flipped out is actually the mechanism by which Rad4 detects the damage. Thus, in this case, the target for the subsequent repair is not directly recognized by Rad4 at all, andinstead the protein uses flipping as an indirect mechanism to detect the loss of a normal base-paired DNA double helix.
When a base is removed from DNA, the reaction is followed by excision of the phosphodiester backbone by an endonuclease, DNA synthesis by a DNA polymerase to fill the gap, and ligation by a ligase to restore the integrity of the polynucleotide chain, as described for the nucleotide excision repair pathways in the previous section.



|
|
|
|
دخلت غرفة فنسيت ماذا تريد من داخلها.. خبير يفسر الحالة
|
|
|
|
|
|
|
ثورة طبية.. ابتكار أصغر جهاز لتنظيم ضربات القلب في العالم
|
|
|
|
|
|
|
العتبة العباسية المقدسة تقدم دعوة إلى كلية مزايا الجامعة للمشاركة في حفل التخرج المركزي الخامس
|
|
|