


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية| Recombining Meiotic Chromosomes Are Connected by the Synaptonemal Complex |
|
|
|
Read More
Date: 3-6-2021
Date: 24-3-2021
Date: 30-11-2015
|
Recombining Meiotic Chromosomes Are Connected by the Synaptonemal Complex
KEY CONCEPTS
- During the early part of meiosis, homologous chromosomes are paired in the synaptonemal complex.
- The mass of chromatin of each homolog is separated from the other by a proteinaceous complex.
A basic paradox in recombination is that the parental chromosomes never seem to be in close enough contact for recombination of DNA to occur. The chromosomes enter meiosis in the form of replicated (sister chromatid) pairs, which are visible as a mass of chromatin.
They pair to form the synaptonemal complex, and it has been assumed for many years that this represents some stage involved with recombination—possibly a necessary preliminary to exchange of DNA. A more recent view is that the synaptonemal complex is a consequence rather than a cause of recombination, but we have yet to define how the structure of the synaptonemal complex relates to molecular contacts between DNA molecules.
Synapsis begins when each chromosome (sister chromatid pair) condenses around a proteinaceous structure called the axial element. The axial elements of corresponding chromosomes then become aligned, and the synaptonemal complex forms as a tripartite structure, in which the axial elements, now called lateral elements, are separated from each other by a central element. FIGURE 1. shows an example.
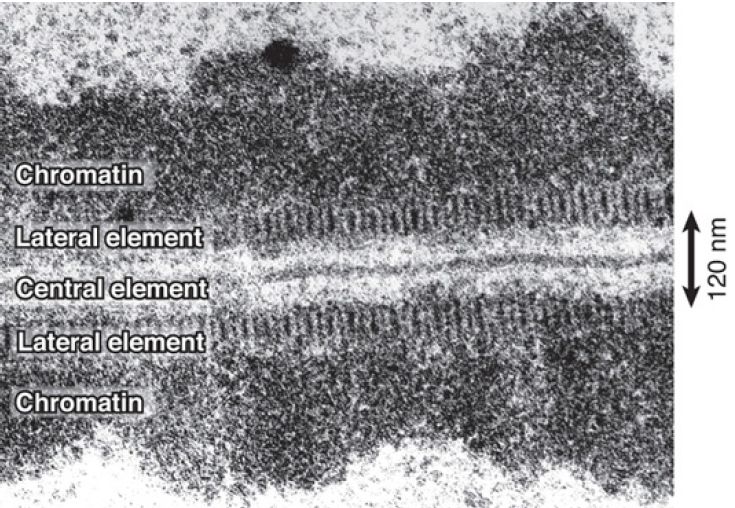
FIGURE 1. The synaptonemal complex brings chromosomes into juxtaposition.
Reproduced from D. von Wettstein. Proc. Natl. Acad. Sci. USA 68 (1971): 851–855. Photo courtesy of Diter von Wettstein, Washington State University.
Each chromosome at this stage appears as a mass of chromatin bounded by a lateral element. The two lateral elements are separated from each other by a fine, but dense, central element.
The triplet of parallel dense strands lies in a single plane that curves and twists along its axis. The distance between the homologous chromosomes is considerable in molecular terms at more than 200 nm (the diameter of DNA is 2 nm). Thus, a major problem in understanding the role of the complex is that, although it aligns homologous chromosomes, it is far from bringing
homologous DNA molecules into contact.
The only visible link between the two sides of the synaptonemal complex is provided by spherical or cylindrical structures observed in fungi and insects. They lie across the complex and are called nodes or recombination nodules; they occur with the same frequency and distribution as the chiasmata. Their name reflects the possibility that they may prove to be the sites of recombination. From mutations that affect synaptonemal complex formation, we can relate the types of proteins that are involved to its structure. FIGURE 2 presents a molecular view of the synaptonemal complex. Its distinctive structural features are due to two groups of proteins:
-The cohesins form a single linear axis for each pair of sister chromatids from which loops of chromatin extend. This is equivalent to the lateral element of Figure 1. (The cohesins belong to a general group of proteins involved in connecting sister chromatids so that they segregate properly at mitosis or meiosis; they are discussed further in the chapter titled Epigenetics II.)
- The lateral elements are connected by transverse filaments that are equivalent to the central element of Figure 2. These are formed from Zip proteins.
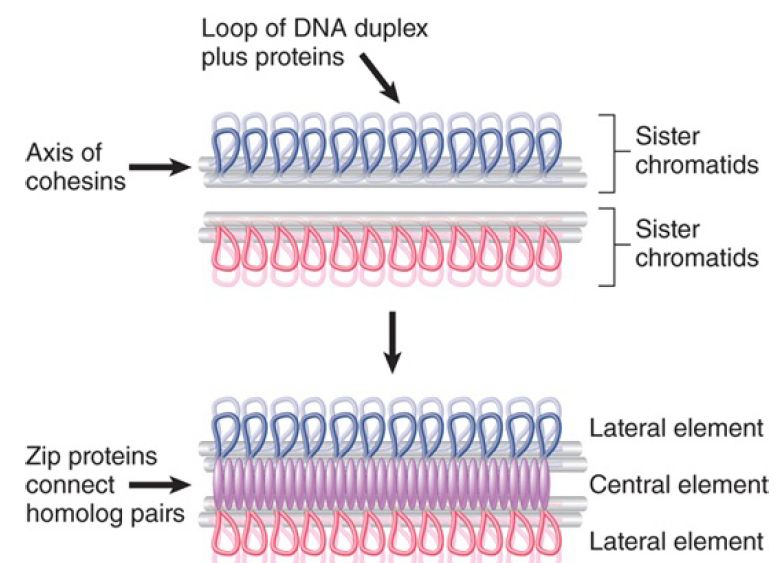
FIGURE 2. Each pair of sister chromatids has an axis made of cohesins. Loops of chromatin project from the axis. The synaptonemal complex is formed by linking together the axes via Zip proteins.
Mutations in proteins that are needed for lateral elements to form are found in the genes coding for cohesins. The cohesins that are used in meiosis include Smc3 (which is also used in mitosis) and Rec8 (which is specific to meiosis and is related to the mitotic cohesin Scc1). The cohesins appear to bind to specific sites along the chromosomes in both mitosis and meiosis. They are likely to play a structural role in chromosome segregation. At meiosis, the formation of the lateral elements may be necessary for the later stages of recombination, because although these mutations do not prevent the formation of DSBs, they do block formation of recombinants.
The zip1 mutation allows lateral elements to form and to become aligned, but they do not become closely synapsed. The N-terminal domain of the Zip1 protein is localized in the central element, but the C-terminal domain is localized in the lateral elements. Two other proteins, Zip2 and Zip3, are also localized with Zip1. The group of Zip proteins forms transverse filaments that connect the lateral elements of the sister chromatid pairs.



|
|
|
|
علامات بسيطة في جسدك قد تنذر بمرض "قاتل"
|
|
|
|
|
|
|
أول صور ثلاثية الأبعاد للغدة الزعترية البشرية
|
|
|
|
|
|
|
قسم الشؤون الفكرية والثقافية يجري اختبارات مسابقة حفظ دعاء أهل الثغور
|
|
|