


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 10-4-2021
Date: 16-11-2020
Date: 23-12-2015
|
Do Nucleosomes Lie at Specific Positions?
KEY CONCEPTS
-Nucleosomes can form at specific positions as the result of either the local structure of DNA or proteins that interact with specific sequences.
-A common cause of nucleosome positioning is when proteins binding to DNA establish a boundary.
-Positioning can affect which regions of DNA are in the linker and which face of DNA is exposed on the nucleosome surface.
-DNA sequence determinants (exclusion or preferential binding) might be responsible for half of the in vivo nucleosome positions.
Does a particular DNA sequence always lie in a certain position in vivo with regard to the topography of the nucleosome? Or, are nucleosomes arranged randomly on DNA so that a particular sequence can occur at any location—for example, in the core region in one copy of the genome and in the linker region in another?
To investigate this question, it is necessary to use a defined sequence of DNA; more precisely, we need to determine the position relative to the nucleosome of a defined point in the DNA. FIGURE 1 illustrates the principle of a procedure used to achieve this.
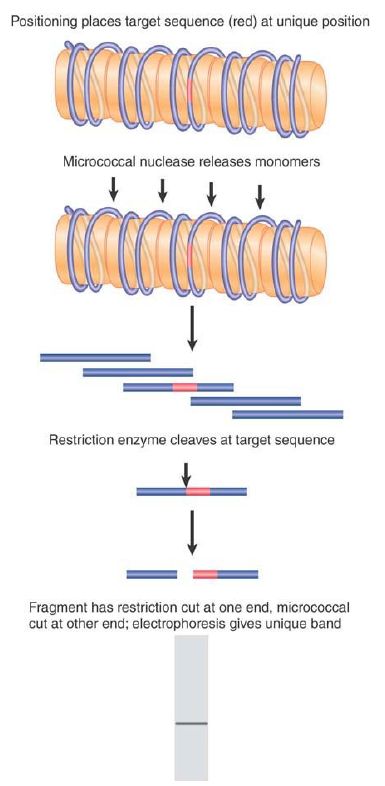
FIGURE 1. Nucleosome positioning places restriction sites at unique positions relative to the linker sites cleaved by micrococcal nuclease.
Suppose that the DNA sequence is organized into nucleosomes in only one particular configuration so that each site on the DNA always is located at a particular position on the nucleosome. This type of organization is called nucleosome positioning (or sometimes nucleosome phasing). In a series of positioned nucleosomes, the linker regions of DNA comprise unique sites.
Consider the consequences for just a single nucleosome. Cleavage with MNase generates a monomeric fragment that constitutes a specific sequence. If the DNA is isolated and cleaved with a restriction enzyme that has only one target site in this fragment, it should be cut at a unique point. This produces two fragments, each of unique size.
Researchers separate the products of the MNase/restriction enzyme double digest by gel electrophoresis. They then use a probe representing the sequence on one side of the restriction site to identify the corresponding fragment in the double digest. This technique is called indirect end labeling (because it is not possible to label the end of the nucleosomal DNA fragment itself, it must be detected indirectly with a probe).
Reversing the argument, the identification of a single sharp band demonstrates that the position of the restriction site is uniquely defined with respect to the end of the nucleosomal DNA (as defined by the MNase cut). Thus, the nucleosome has a unique sequence of DNA. If a given region contains an array of positioned nucleosomes, researchers can map the position of each by using this method. FIGURE 2 shows an example of a gene promoter containing an ordered array of nucleosomes. In this MNase map, numerous positioned nucleosomes can be identified, indicated by the ovals to the left. Note that the TATA box is covered by a nucleosome; in this example this gene is not transcriptionally active.
What happens if the nucleosomes do not lie at a single position? Now the linkers consist of different DNA sequences in each copy of the genome. Thus, the restriction site lies at a different position each time; in fact, it lies at all possible locations relative to the ends of the monomeric nucleosomal DNA. FIGURE 3 shows that the double cleavage then generates a broad smear, ranging from the smallest detectable fragment (~20 bases) to the length of the monomeric DNA. Although the indirect end-labeling method is appropriate for monitoring nucleosome positioning at individual loci, MNase digestion can also be combined with massively parallel DNA sequencing to define nucleosome locations on a genome-wide scale.
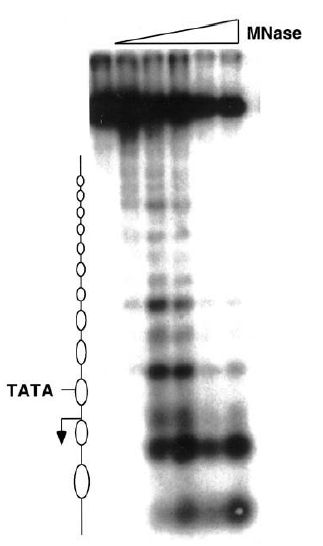
FIGURE 2 An MNase map of nucleosome positions in an inactive gene. The lanes from left to right have been treated with increasing amounts of MNase. The nucleosomes occupy the regions that lack cut sites (indicated by ovals) and are arranged in a well-ordered array. The position of the TATA box and the transcriptional start site (arrow) are indicated.
Figure courtesy of Dr. Jocelyn Krebs.
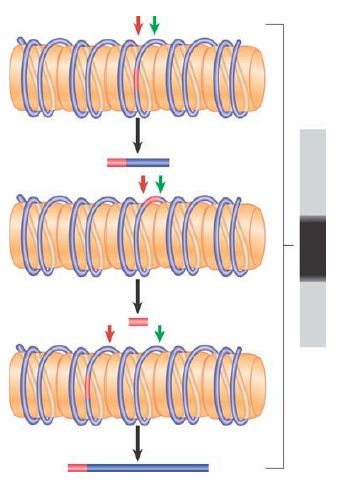
FIGURE 3 In the absence of nucleosome positioning, a restriction site can lie at any possible location in different copies of the genome. Fragments of all possible sizes are produced when a restriction enzyme cuts at a target site (red) and micrococcal nuclease cuts at the junctions between nucleosomes (green).
In discussing these experiments, we have treated MNase as an enzyme that cleaves DNA at the exposed linker regions without any sort of sequence specificity. MNase does have some sequence specificity, though, which is biased toward selection of A-T–rich sequences. Thus, we cannot assume that the existence of a specific band in the indirect end-labeling technique represents the distance from a restriction cut to the linker region. It could instead represent the distance from the restriction cut to a preferred micrococcal nuclease cleavage site.
This possibility is controlled by treating the naked DNA in exactly the same way as the chromatin. If there are preferred sites for MNase in the particular region, specific bands are found.
Researchers can compare this pattern of bands with the pattern generated from chromatin.
A difference between the control DNA band pattern and the chromatin pattern provides evidence for nucleosome positioning.
Some of the bands present in the control DNA digest might disappear from the nucleosome digest, indicating that preferentially cleaved positions are unavailable. New bands might appear in the nucleosome digest when new sites are rendered preferentially accessible by the nucleosomal organization.
Nucleosome positioning might be accomplished in either of two ways:
- Intrinsic mechanisms: Nucleosomes are deposited specifically at particular DNA sequences, or are excluded by specific sequences. This modifies our view of the nucleosome as a subunit able to form between any sequence of DNA and a histone octamer.
- Extrinsic mechanisms: The first nucleosome in a region is preferentially assembled at a particular site due to action of other protein(s). A preferential starting point for nucleosome positioning can result either from the exclusion of a nucleosome from a particular region (due to competition with another protein binding that region), or by specific deposition of a nucleosome at a particular site. The excluded region of the positioned nucleosome provides a boundary that restricts the positions available to the adjacent nucleosome. A series of nucleosomes can then be assembled sequentially, with a defined repeat length.
We know that the deposition of histone octamers on DNA is not random with regard to sequence. The pattern is intrinsic in cases in which it is determined by structural features in DNA. It is extrinsic in other cases, resulting from the interactions of other proteins with the DNA and/or histones.
Certain structural features of DNA affect placement of histone octamers. DNA has intrinsic tendencies to bend in one direction rather than another. For example, AT dinucleotides bend easily, and thus A-T–rich sequences are easier to wrap tightly in a nucleosome. A-T–rich regions locate so that the minor groove faces in toward the octamer, whereas G-C–rich regions are arranged so that the minor groove points outward. Long runs of dA-dT (>8 bp), in contrast, stiffen the DNA and avoid positioning in the central, tight, superhelical turn of the core. It is not yet possible to sum all of the relevant structural effects and thus entirely predict the location of a particular DNA sequence with regard to the nucleosome, although recently researchers have developed some predictive models that appear to match at least some in vivo positioning data. Sequences that cause DNA to take up more extreme structures have effects such as the exclusion of nucleosomes, and thus cause boundary effects or nucleosome-free regions.
Positioning of nucleosomes near boundaries is common. If there is some variability in the construction of nucleosomes—for example, if the length of the linker can vary by, say, 10 bp—the specificity of positioning would decline proceeding away from the first, defined nucleosome at the boundary. In this case, we might expect the positioning to be maintained rigorously only relatively near the boundary.
The location of DNA on nucleosomes can be described in two ways. FIGURE 4 shows that translational positioning describes the position of DNA with regard to the boundaries of the nucleosome. In particular, it determines which sequences are found in the linker regions. Shifting the DNA by 10 bp brings the next turn into a linker region. Thus, translational positioning determines which regions are more accessible (at least as judged by sensitivity to MNase).
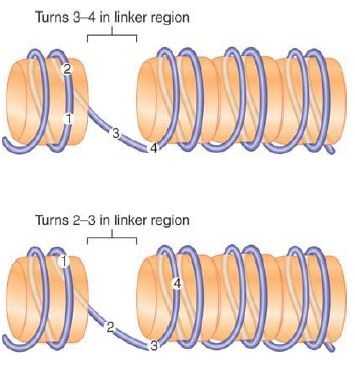
FIGURE 4. Translational positioning describes the linear position of DNA relative to the histone octamer. Displacement of the DNA by 10 bp changes the sequences that are in the more exposed linker regions, but does not necessarily alter which face of DNA is protected by the histone surface and which is exposed to the exterior.
DNA lies on the outside of the histone octamer. As a result, one face of any particular sequence is obscured by the histones, whereas the other face is exposed on the surface of the nucleosome. Depending upon its positioning with regard to the nucleosome, a site in DNA that must be recognized by a regulatory protein could be inaccessible or available. The exact position of the histone octamer with respect to DNA sequence can therefore be important. FIGURE 5 shows the effect of rotational positioning of the double helix with regard to the octamer surface.
If the DNA is moved by a partial number of turns (imagine the DNA as rotating relative to the protein surface), there is a change in the exposure of sequence to the outside.
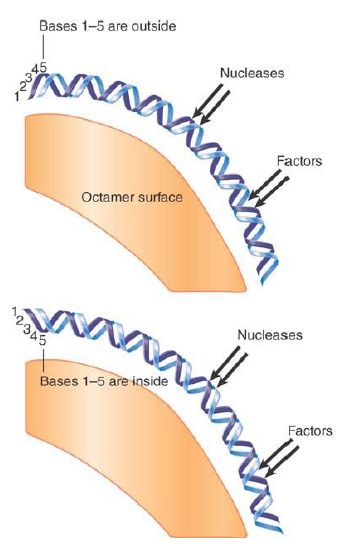
FIGURE 5. Rotational positioning describes the exposure of DNA on the surface of the nucleosome. Any movement that differs from the helical repeat (~10.2 bp/turn) displaces DNA with reference to the histone surface. Nucleotides on the inside are more protected against nucleases than nucleotides on the outside.
Both translational and rotational positioning can be important in controlling access to DNA. The best characterized cases of positioning involve the specific placement of nucleosomes at promoters. Translational positioning and/or the exclusion of nucleosomes from a particular sequence might be necessary to allow a transcription complex to form. Some regulatory factors can bind to DNA only if a nucleosome is excluded to make the DNA freely accessible, and this creates a boundary for translational positioning. In other cases, regulatory factors can bind to DNA on the surface of the nucleosome, but rotational positioning is important to ensure that the face of DNA with the appropriate contact points is exposed.
We discuss the connection between nucleosomal organization and transcription in the chapter titled Eukaryotic Transcription Regulation, but note for now that promoters (and some other structures) often have short regions that exclude nucleosomes.
These regions typically form a boundary next to which nucleosome positions are restricted. A survey of an extensive region in the Saccharomyces cerevisiae genome (mapping 2,278 nucleosomes over 482 kb of DNA) showed that in fact 60% of the nucleosomes have specific positions as the result of boundary effects, most often from promoters. Nucleosome positioning is a complex output of intrinsic and extrinsic positioning mechanisms. Thus, it has been difficult to predict nucleosome positioning based on sequence alone, though there have been some successes. Large-scale sequencing studies of isolated nucleosomal DNA have revealed intriguing sequence patterns found in positioned nucleosomes in vivo, and it is estimated that 50% or more of in vivo nucleosome positioning is the result of intrinsic sequence determinants encoded in the genomic DNA. It is also important to note that even when a dominant nucleosome position is detected experimentally, it is not likely to be completely invariant (i.e., the nucleosome is not in that exact position in every cell in a sample); instead, it represents the
most common location for a nucleosome in that region out of larger set of related positions.



|
|
|
|
تفوقت في الاختبار على الجميع.. فاكهة "خارقة" في عالم التغذية
|
|
|
|
|
|
|
أمين عام أوبك: النفط الخام والغاز الطبيعي "هبة من الله"
|
|
|
|
|
|
|
قسم شؤون المعارف ينظم دورة عن آليات عمل الفهارس الفنية للموسوعات والكتب لملاكاته
|
|
|