
The nitrogen cycle
 المؤلف:
Peter Atkins, Tina Overton, Jonathan Rourke, Mark Weller, and Fraser Armstrong
المؤلف:
Peter Atkins, Tina Overton, Jonathan Rourke, Mark Weller, and Fraser Armstrong
 المصدر:
Shriver and Atkins Inorganic Chemistry ,5th E
المصدر:
Shriver and Atkins Inorganic Chemistry ,5th E
 الجزء والصفحة:
ص765-768
الجزء والصفحة:
ص765-768
 2025-10-26
2025-10-26
 52
52
The nitrogen cycle
Key points: The nitrogen cycle involves enzymes containing Fe, Cu, and Mo, often in cofactors having very unusual structures; nitrogenase contains three different kinds of FeS cluster, one of which also contains Mo and a small interstitial atom. The global biological nitrogen cycle involves organisms of all types and a diverse variety of metalloenzymes (Fig. 27.46 and Box 15.2). The cycle can be divided into uptake of us able nitrogen (assimilation) from nitrate or N2 and denitrification (dissimilation). The nitrogen cycle involves many different organisms and a variety of metal-containing enzymes.
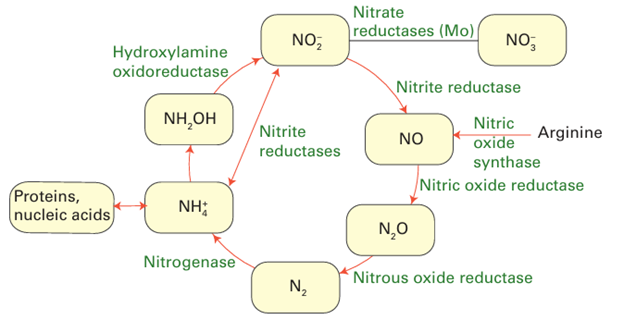
Figure 27.46 The biological nitrogen cycle.
Many of the compounds are toxic or environmentally challenging. Ammonia is a crucial compound for the biosynthesis of amino acids and NO3 is used as an oxidant. Molecules such as NO are produced in small amounts to serve as cell signalling agents that play a crucial in physiology and health. Nitrous oxide, which is isoelectronic with CO2, is a potential greenhouse gas: its release to the atmosphere depends on the balance between activities and abundancies of NO reductase and N2O reductase across the bio logical world. The so-called ‘nitrogen-fixing’ bacteria found in soil and root nodules of certain plants contain an enzyme called nitrogenase that catalyses the reduction of N2 to ammonia in a reaction that is coupled to the hydrolysis of 16 molecules of ATP and the production of H2:
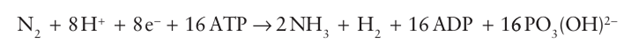
Fixed’ nitrogen is essential for the synthesis of amino acids and nucleic acids, so it is central to agricultural production. Industrial production of ammonia by the Haber process (Section 26.12) involves reaction of N2 and H2 at high pressures and high temperatures; by contrast, nitrogenase produces NH3 under normal conditions, and it is small wonder that it has attracted so much attention. Indeed, the mechanism of activation of the N2 molecule by nitrogenase has inspired coordination chemists for several decades. The process is very costly in terms of energy for a reaction that is not very unfavourable thermodynamically. However, as we saw in Section 15.6, N2 is an unreactive molecule and energy is required to overcome the high activation barrier for its reduction. Nitrogenase is a complex enzyme that consists of two types of protein: the larger of the two is called the ‘MoFe-protein’ and the smaller is the ‘Fe-protein’ (Fig. 27.47). The Fe-protein contains a single [4Fe–4S] cluster that is coordinated by two cysteine residues from each of its two subunits. The role of the Fe-protein is to transfer electrons to the MoFe-protein in a reaction that is far from understood: in particular, it is unclear why each electron transfer is accompanied by the hydrolysis of two ATP molecules, which are bound to the Fe-protein.
The MoFe protein is α2 β2, each α pair of which contains two types of supercluster. The [8Fe 7S] cluster (49) is known as the ‘P-cluster’ and is thought to be an electron transfer centre, whereas the other cluster (50), formulated as [Mo7Fe 8S, X] and known as ‘FeMoco’ (FeMo cofactor), is thought to be the site at which N2 is reduced to NH3. The Mo is coordinated also by an imidazole-N from histidine and two O atoms from an exogenous molecule R-homocitrate. The mechanism of N2 reduction is still unresolved, the main question being whether N2 is bound and reduced at the Mo atom or at some other site. Here it is significant that nitrogenases are known in which the Mo atom is replaced by a V or Fe atom, arguing against a specific role for Mo. The cage-like cluster provided by the six central Fe atoms also coordinates a small central atom (‘X’),
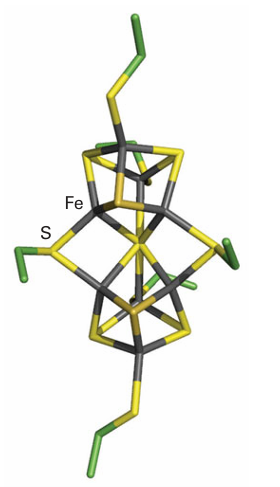
49 Nitrogenase P-cluster [8Fe-7S]
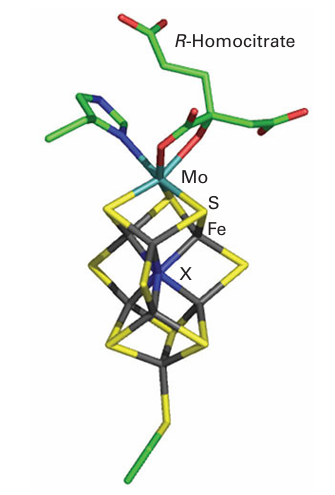
50 Nitrogenase FeMoco [Mo7Fe–8S,X]
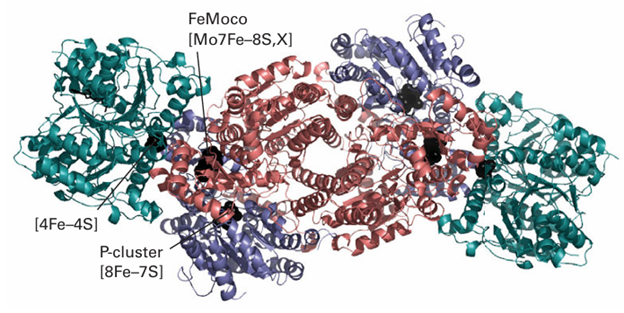
Figure 27.47 The structure of nitrogenase showing the Fe protein and the MoFe protein complexed with each other. Positions of the metal centres (black) are indicated. The MoFe protein is an α2β2 of different subunits (red and blue) and it contains two P-clusters and two MoFe cofactors. The Fe protein (green) has a [4Fe 4S] cluster and it is also the site of binding and hydrolysis of Mg-ATP.
which is proposed to be C, N, or O. It was nearly ten years after the determination of the structure of the MoFeS cage that this small central atom was discovered, after improve ments in resolution and detailed consideration of the X-ray interference characteristics. The six Fe atoms would otherwise be three-coordinate with a flattened trigonal-pyramidal geometry. Nitrate reductase is another example of a Mo enzyme involved in the transfer of an O atom, in this case catalysing a reduction reaction (the standard potential for the NO3/NO2 couple corrected to pH=7 is+0.4 V, thus NO3 is quite strongly oxidizing). The other enzymes in the nitrogen cycle contain either haem or Cu as their active sites. There are two distinct classes of nitrite reductase. One is a multi-haem enzyme that can reduce nitrite all the way to NH3. The other class contains Cu and carries out one-electron transfer, producing NO: it is a trimer of identical subunits, each of which contains one ‘blue’ Cu (mediating long-range electron transfer to the electron donor, usually a small ‘blue’ Cu protein) and a Cu centre with more conventional tetragonal geometry that is thought to be the site of nitrite binding.
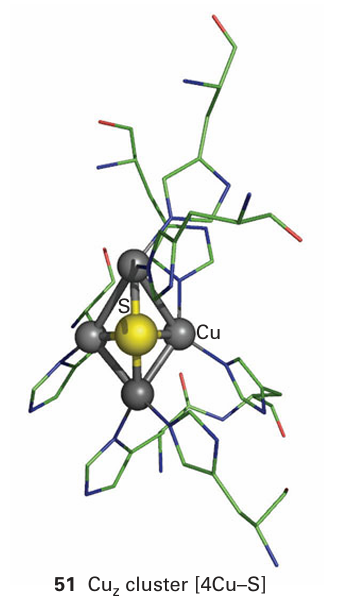
The nitrogen cycle is notable for using some of the most unusual redox centres yet encountered as well as some of the strangest reactions. Another unusual cofactor is a [4Cu S] cluster, named CuZ , that is found in N2 O reductase and has the structure shown in (51). It is puzzling how this centre is able to bind and activate N2 O, which is a poor ligand. Long-range electron transfer in N2 O reductase is carried out by a CuA centre, the same as found for cytochrome c oxidase. Of particular importance for humans are two enzymes that manipulate NO. One of these, NO synthase, is a haem enzyme responsible for producing NO, by oxidation of l-arginine, on receipt of a signal. Its activity is controlled by calmodulin (Section 27.4). The other enzyme is guanylyl cyclase, which catalyses the formation of the important regulator cyclic guanosine monophosphate (cGMP) from guanosine triphosphate. Nitric oxide binds to the haem Fe of guanylyl cyclase, displacing a histidine ligand and activating the enzyme. Another interesting NO-binding protein is nitrophorin, which is found in some bloodsucking parasites, notably ‘kissing bugs’ (predacious bugs of the family Reduviidae). Nitrophorin binds NO tightly until it is injected into a victim, where a change in pH causes its release. The free NO causes dilation of the surrounding blood vessels, rendering the victim a more effective blood donor.
 الاكثر قراءة في مواضيع عامة في الكيمياء العضوية
الاكثر قراءة في مواضيع عامة في الكيمياء العضوية
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


