


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 28-11-2015
Date: 6-12-2015
Date: 16-5-2016
|
Histones
Histones are small basic (positively charged) proteins that are complexed with DNA in the nucleus of almost all eukaryotic cells (not dinoflagellates or mammalian spermatozoa) and serve to package the DNA into nucleosomes in chromatin. It has recently become clear that they also play an important regulatory role in gene transcription and repression. Together, the histones occur in a mass roughly equal to that of DNA. There are five histone types, falling into two functional classes: the core histones (H3, H4, H2A, and H2B) and the larger linker histones (H1 and its variants). Two copies of each of the core histones form the histone octamer that constitutes the protein core of the nucleosome, around which are wound two complete left-handed superhelical turns of DNA. The nucleosome is stabilized by the binding of one molecule of the histone H1 class, which also plays a role in stabilization of the higher-order structure of chromatin. It is becoming clear that H1 may also play a specific role in the regulation of some genes. Both the core histones and H1 undergo dynamic reversible modification. Acetylation of the core histones plays an important role in processes as diverse as transcription, nucleosome assembly, and heterochromatin formation. Histone H1 is phosphorylated during S-phase and mitosis in the cell cycle, and its extreme cell-type specific variants in avian erythrocytes and sea urchin sperm are phosphorylated up until the final stages of chromatin compaction. Two volumes contain a wealth of information about the histones (1, 2).
1. Core Histones: The Histone Octamer
The core histones [molecular masses ~11,300Da (H4) to 15,300 (H3)] show a high degree of evolutionary sequence conservation, consistent with their role as organizing structural components in a common mode of DNA packaging in eukaryotes. Histones H3 and H4 are the most conserved proteins known, with only two (conservative) amino acid differences out of 102 residues between pea and calf H4. All the core histones have high contents of the basic amino acids lysine and arginine (whose side chains are protonated, and hence positively charged, at neutral pH). H2A and
H2B are relatively lysine-rich, and H3 and H4 are relatively arginine-rich. The basic residues are unevenly distributed along the amino acid sequence and are concentrated toward the N-terminal regions of about 25 to 40 residues. These regions (the N-terminal “tails”) are implicated in stabilization of chromatin higher-order structure and are essential for a number of vital functions. They are also the sites of several post-translational modifications, including acetylation, which has been well-studied.
The four core histones exist in chromatin as an octamer that is structurally organized as a tetramer of H3 and H4 (H32H42) and two dimers of H2A and H2B (H2A.H2B). The octamer may be isolated as an entity from chromatin at high ionic strength (eg, 2 M NaCl), which disrupts the electrostatic interactions between the histones and DNA and, in addition, preserves the association of tetramers and dimers (3). At physiological ionic strength, in the absence of DNA, the isolated octamer falls apart into tetramers and dimers. The structure of the octamer, determined to 3.1 Å resolution by X-ray crystallography (4), reveals two distinctive features of the histone associations (Fig. 1). First, the four histones (which have no obvious sequence homology) share a common fold, now designated the “histone fold.” This comprises a long central a-helix and two shorter flanking a-helices connected by loops. Second, the histone pairs H3, H4 and H2A, H2B share a common mode of association as heterodimers in a “handshake motif,” creating a crescent-shaped structure. Two H3.H4 heterodimers interact through a-helical regions on H3 to form the rather flat tetramer, which resembles a twisted horseshoe; H2A.H2B dimers flank this on each face, through interactions between a-helical regions on H2B and H4. The octamer crystal structure confirmed the presence of a left-handed helical ramp)for DNA binding) on the octamer surface, which had been inferred from earlier work (5), and suggested how the histone fold might form DNA contacts. The details of the interaction of the octamer with DNA have since been observed directly in the high-resolution structure of the nucleosome core particle, which also reveals the positions of the histone tails that were not visible in the structure of the octamer alone, because they are disordered. Intriguingly, the histone fold has since been found embedded in much larger proteins, notably several TAFs (TATA-box binding protein-associated factors) and transcription factors.
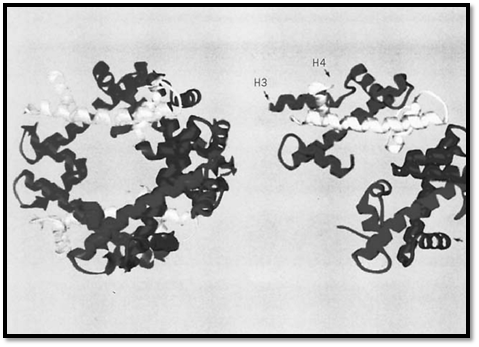
Figure 1. The structure of the histone octamer at 3.1 Å resolution, showing the histone folds (4).
The N-terminal histone tails of the core histones are well-conserved, despite being external to the structured core of the octamer, suggesting some structural or functional role. The tails do not appear to be required for nucleosome core integrity and may be removed by proteolysis without causing structural disruption. There is good evidence that they play a role, together with the basic tails of H1, in stabilization of chromatin higher-order structure. In the structure of the nucleosome core particle (6) they form few, if any, contacts within the nucleosome core, but are extended from it and are well-placed to interact with neighboring nucleosomes or other chromosomal components. The role of the tails has been extensively studied genetically in yeast (7). It is increasingly recognized that the tails may function as recognition sites for protein factors important for chromatin function. For example, particular sequences in the N-terminal tails of H4 (residues 16 to 29) and H3 (residues 4 to 20) are required, together with other proteins (Rap1, Sir2, Sir3, and Sir4 ) for transcriptional silencing (through formation of a more repressed chromatin structure) at yeast telomeres and silent mating type loci. The amino-termini of H3 and H4 are also required for a1–a2 repression in yeast (a regulatory mechanism involved in distinguishing diploid from haploid yeast cells) (8) and for the action in vitro of at least one chromatin remodeling machine, Drosophila NURF (9).
The N-terminal tails are the sites of several types of post-translational modification, the best studied of which is acetylation. It is involved in transcription and chromatin assembly, the acetylation patterns being different in the two cases, and could also provide additional markers for specific recognition by various partner proteins. The specific acetylation of Lys12 only in heterochromatin in Drosophila and yeast could perform such a role. Transcription-linked hyperacetylation, primarily on H4 and H3, is believed to result in relaxation of the repressive higher-order structure by disruption of internucleosome contacts. Crystal contacts in the nucleosome core particle suggest contacts that might be involved (6). Other post-translational modifications have been less well studied and are less well understood (1, 2). Phosphorylation occurs constitutively at the N-terminal serine of H2A and, in a small proportion of histones, at Ser 10 and Ser 28 of H3 in response to mitogen stimulation in mammalian cells (10). Phosphorylation and acetylation appear to coexist in the same nucleosomes after mitogen stimulation and might act synergistically to disrupt internuclesomal contacts involving H3 tails and to facilitate transcription. The role of other modifications of the core histones (methylation of lysine side chains, and—in the case of H2B—ADP-ribosylation ) is unclear; ADP-ribosylation may have a role in DNA repair, probably by disruption of chromatin structure. Also unclear is the role of ubiquitination of lysine side chains near the C-terminus of H2A and H2B (ie, covalent attachment of the small protein ubiquitin through an isopeptide linkage), but it appears not to be related to the well-recognized role of ubiquitination as a signal for protein degradation.
Core histone variants, the products of distinct genes, are found in all organisms. Their precise role is not clear, but they presumably allow fine-tuning of nucleosome structure and stability for particular purposes. A well-documented example is the developmental pattern of expression of five H2A variants and four H2A variants, which changes through the cleavage, blastula, and gastrula stages of sea urchin embryogenesis (11). Significantly, there is no variation in H3 and H4, which form the structural core of the nucleosome. During spermatogenesis in the same organism, there is a global replacement of somatic H2B with the larger and more basic sperm-specific variant spH2B, which has a long N-terminal extension with several “-SPKK-” (-Ser/Thr-Pro-X/Lys-Lys/Arg-) motifs and helps in the tight compaction of chromatin in the sperm head. On fertilization, this is phosphorylated and then replaced during subsequent cell divisions with somatic variants. Drosophila has two evolutionarily conserved H2A variants: H2A.X, which has a C-terminal extension (like wheat H2A1, which has 19 extra residues and binds to linker DNA ), and H2A.vD, which is essential for Drosophila development and has counterparts in mammals (H2A.Z), chickens (H2A.F/Z), and the ciliated protozoan Tetrahymena (hv1), where it is found in actively transcribed chromatin. H2A.Z is interesting because it has an N-terminal tail resembling that of H4 and indeed is acetylated more than the canonical H2A, thus providing obvious additional possibilities for regulation of function. A class of more extreme so-called core histone variants are really hybrid proteins, with a region of histone sequence fused to a wholly unrelated region of unknown function. Two such proteins are the mammalian centromere-specific protein CENP-A (the yeast homologue is CSE4) and macroH2A. CENP-A (molecular mass ~17,000Da) is similar to H3 in its C-terminal domain, but has a highly divergent N-terminal region and appears to be associated, although not solely, with a- satellite DNA (12) . Assuming that it replaces the normal H3, it presumably imparts special properties to centromeric chromatin. MacroH2A is a highly conserved protein in which the N-terminal third resembles H2A and the C-terminal two-thirds contains a coiled-coil protein dimerization motif. There are two macroH2A subtypes, which are (a) highly conserved and (b) identical in the histone region. It came as a surprise to find that one subtype is localized to the inactive X-chromosome (in mouse) and is distributed throughout the chromatin (13); it is tempting to speculate that the coiled-coil might have a role in protein oligomerization and condensation of chromatin. MacroH2A is thus one more distinctive component of the inactive X-chromosome, the others being heavily methylated DNA, hypoacetylated H4, and association with a large cis-acting nuclear RNA (termed Xist. (
2. Linker Histones/Histone H1
Linker histones (H1 and its variants and subtypes) in higher eukaryotes are larger than the core histones (molecular mass ~20,000 to 25,000 Da) and are particularly lysine-rich. They have a tripartite domain structure, in which a central globular domain of about 80 amino acid residues, which is highly conserved between species, is flanked by basic N- and C-terminal domains (“tails”) that are much more divergent. In the absence of DNA, the tails are disordered and may be selectively removed by proteolysis, leaving the globular domain intact. In a typical mammalian H1, the N- and C-terminal tails are about 40 and 100 residues long, respectively; the lengths differ in some species-specific and cell-type-specific variants. Nucleosomes released from chromatin by digestion with micrococcal nuclease (Staphylococcal nuclease) are rapidly trimmed by further digestion so that the ~200 bp of DNA is reduced to ~166 bp, giving a chromatosome. If H1 is removed before digestion, however, only 146 bp remain protected, as digestion proceeds to the limit nucleosome core particle. The globular domain of H1 alone is sufficient to protect 166 bp, so this is the region of H1 that binds close to the nucleosome core, stabilizing the nucleosome and protecting an extra 20 bp from digestion. The basic C-terminal tail (and possibly also the N-terminal tail) binds to the linker DNA between adjacent nucleosomes (hence the name linker histones), partially neutralizing its negative charge and promoting folding of the nucleosome filament into a higher-order structure, the 30-nm filament; H1 is located on the inside of the 30-nm filament. The role of H1 is thus to stabilize both the nucleosome and an ordered higher-order chromatin structure. H1 may be phosphorylated and poly-ADP-ribosylated; the role of the latter modification is not clear, but it is probably involved in DNA repair .
There are many subtypes and more extreme sequence variants of H1, which all share the same domain organization. These may coexist in the same cell type (eg, there are six H1 subtypes in chicken erythrocytes), and the assumption is that they might stabilize different chromatin higher-order structures. This would be likely to occur only if they were clustered along a chromatin filament, and this indeed appears to be the case for one of the seven H1 variants in the interphase polytene chromosomes in the Dipteran insect Chironomus thummi (14). Information is lacking for other systems or for other H1s in Chironomus. There are several instances of changes in linker histone variants during developmentally regulated processes (11, 15). During sea urchin embryogenesis, six distinct H1 subtypes are produced at particular stages; and Xenopus has an embryonic form of H1, namely B4, that is very different from the normal somatic H1. The C-terminal tail is less basic, and this might result in a less condensed chromatin structure in Xenopus embryos, compatible with rapid nuclear division and DNA replication. Extreme variants may also be produced to shut down transcription in particular cell types, and they appear to be associated with more stable chromatin higher-order structures. For example, in transcriptionally inert mature avian erythrocytes, H5 (which binds more tightly than H1, probably due to its higher arginine content, although it has shorter N- and C-terminal tails) has largely replaced H1; and in sea urchin sperm, somatic H1 is replaced by a larger and more basic sperm-specific H1, namely spH1, which, together with sperm-specific core histones, is very effective in condensation of the DNA. (In mammalian sperm, histones are replaced by protamines, small arginine-rich proteins, rather than by specialized histone variants.) H1 0, an H1 variant whose appearance in mammalian cells appears to correlate with terminal differentiation (eg, in neurons), is much more similar to H5 than to H1 in its globular domain.
The globular domains of histones H1 and H5 (GH1 and GH5) can bind to the body of the nucleosome and protect an additional 20 bp beyond the core particle length from digestion. The structure of GH5, determined by X-ray crystallography, shows a “winged helix” motif (a variant on the helix–turn–helix DNA-binding motif) which is also found in two proteins that bind to specific DNA sequences, namely the liver transcription factor HNF3g (hepatocyte nuclear factor 3g,) and the prokaryotic protein cyclic AMP receptor protein (CRP) (16). It consists of a three-helix bundle with a b-hairpin (or “wing”) at its C-terminus. Based on the X-ray crystal structures of these proteins bound to DNA, the likely mode of binding of GH5 to DNA, in the major groove, has been deduced. Biochemical evidence points to a second DNA binding site on GH5, and this has been identified on the opposite face of the globular domain (17) (Fig. 2). It is likely that the two sites are occupied in the nucleosome by two of the three duplexes (the central turn and the entering and exiting DNA) that are present in the vicinity of the dyad axis of the nucleosome. Current evidence for bulk nucleosomes (18) suggests that GH5 bridges the central turn, close to the dyad, and one of the entering/exiting duplexes. The orientation proposed would place the C-terminal tail of H1 directed toward linker DNA, as expected from various lines of evidence. A different mode of binding of the globular domain to the reconstituted Xenopus 5 S nucleosome has been proposed, but it is not yet clear whether this is a special property of the 5 S nucleosome. The existence of two binding sites on the globular domain is probably the basis of its preference (over linear DNA) for four-way DNA junctions (19), which mimic a pair of duplexes at the DNA crossover point near the nucleosome dyad.
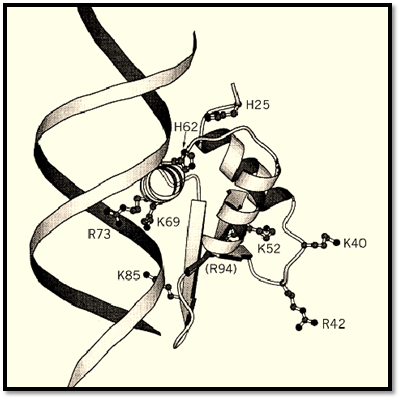
Figure 2. The structure of the globular domain of histone H5 (GH5) showing the two clusters of basic residues at the two proposed DNA-binding sites on opposite faces of the domain (17). Binding to DNA at one site is by analogy with the structures of HNF3g and CRP.
There is no high-resolution structural information for the N- and C-terminal tails of H1 or any of its variants. The role of the N-terminal tail (~40 amino acid residues in the canonical H1) is unclear, but there is some evidence that it may anchor the globular domain correctly in the presence of the C-terminal tail. The basic C-terminal tails (~90 to 130 amino acid residues, depending on the variant( have unusually high contents (~50%) of the basic residues lysine and arginine, as well as relatively high contents of alanine and proline. They are essential for chromatin condensation and are believed to bind to linker DNA. They are disordered in the free protein, but they are likely in the presence of DNA to exist as a-helical segments rich in lysine and alanine, the segments being separated by proline residues (20). The proline residues are often embedded in so-called “-SPKK-” sequence motifs (-Ser/Thr-Pro-X/Lys-Lys/Arg-), where the serine is a potential phosphorylation site. There is some phosphorylation at S-phase of the cell cycle and considerably more at mitosis, probably due to the action of the cyclin-dependent kinase, p34cdc2 (21). The role of phosphorylation is likely to be to loosen the interactions between the H1 tails and linker DNA, and at mitosis to permit other factors and interactions to drive chromosome condensation. Some members of the repertoire of H1s produced during different stages of sea urchin embryogenesis lack -SPKK- motifs, suggesting that the binding of different H1s may be differentially regulated. Phosphorylation also plays a role in the control of chromatin transitions unrelated to the cell cycle, namely, in the late stages of avianerythropoiesis, or sea urchin sperm spermatogenesis, where there is no cell division and where special histone variants that bind more tightly than somatic H1 (ie, H5 and spH1, respectively( suppress transcription by promoting chromatin condensation. H5 and spH1 are phosphorylated at the -SPKK- motifs (of which there are six copies in the N-terminal tail of spH1 and further copies in the C-tail) until the final stage of maturation, when a final dephosphorylation step in the spermatozoon or mature erythrocyte results in the final stage of chromatin condensation and even closer chromatin packing in the sperm head or erythrocyte nucleus; removal of the phosphate increases the net positive charge on the histone and promotes histone–DNA interactions, leading to condensation. An analogous situation exists in the amitotic nucleus of Tetrahymena (22).
There are two very atypical H1s, or—in one case—candidate H1, in lower eukaryotes, which lack the characteristic domain organization of the canonical H1. The ciliated protozoan Tetrahymena thermophila, which has a transcriptionally active but amitotic macronucleus and a transcriptionally inert mitotic micronucleus, contains a small, basic, macronuclear protein of 163 residues, designated H1, that appears to share some of the functions of linker histones. (A set of distinctive polypeptides appears to substitute for H1 in the micronucleus.) The macronuclear H1 condenses chromatin in the nucleus, and phosphorylation at ‘TPVK’ sites results in an increase in nuclear volume, suggesting that phosphorylation loosens interactions with chromatin, as expected (22, 23). It lacks the distinctive globular domain of the canonical H1, however, although its C-terminal region has similarities to the C-terminal tail of H1. Exactly how the Tetrahymena H1 is bound to chromatin is unclear. An unusual, but quite different, domain organization also occurs in a candidate H1 identified from the recently determined complete sequence of the yeast (Saccharomyces cerevisiae( genome. Previous attempts to isolate H1 from yeast had been unsuccessful, and one view was that because the linker length of yeast chromatin was virtually zero (chromatin repeat length ~166 base pairs) there was no requirement for neutralization of linker DNA charge or, therefore, for a canonical linker histone. Analysis of the yeast genome revealed an open reading frame (the HHO1 gene( encoding a protein (Hho1p) with regions of sequence homology to the globular domain of H1, which is regarded as a candidate H1 (24). It has two globular domains of about 80 residues, with a basic N -terminal extension and a basic connecting linker with some resemblance to the C-terminal tail of the canonical H1. Recombinant Hho1p appears to have some of the distinctive properties of H1 in a standard chromatin reconstitution, namely protection of an additional ~20bp of DNA beyond the core particle length against exonuclease digestion (25). Its unique structure, however, means that there are likely to be differences in its detailed mode of binding to chromatin compared with canonical H1, and it is by no means clear at present whether Hho1p functions as a true H1 or is instead a transcription factor with domains homologous to the globular domain of H1 [cf. some TAFs, which have sequence homology to core histones; or even HNF3g which contains the same structural fold as in the globular domains of H5 and H1, although in that case there is no sequence homology between the two proteins].
Gene disruptions and deletions have been used to ask whether H1 is essential. Disruption )“knockout”) of the H10 gene in mouse appeared to be without consequence; it is likely, however, that one or some of the six other subtypes compensate (26); in other words, there is functional redundancy. Deletion of the entire complement of somatic H1 genes has not been achieved. Early Xenopus embryos contain the unusual H1 variant B4 which is later replaced by somatic H1; elimination of B4 had little effect on nuclear assembly or on the development of the organism (27). Deletion of the single gene for the atypical macronuclear H1 of Tetrahymena was not lethal, but was not without effect either (23). Vegetative growth, general transcription, and general nucleosome repeat length were unaffected, but there were changes in the nuclear volume )presumably consistent with the role of the protein in chromatin condensation and packing in the nucleus), in the efficiency of meiotic division, and, significantly, in the transcriptional regulation of specific genes. However, although some genes were activated, consistent with the expected repressive role for H1, others were repressed (eg, the CyP gene, which encodes a thiol proteinase. (One explanation would be that the binding of the Tetrahymena H1 is needed in some nucleosomes to position them in such a way that certain sequences necessary for the binding of activators are exposed. In yeast, deletion of the yeast HHO1 gene had no detectable effect on cell growth, viability, or mating, or on telomeric silencing, basal transcriptional repression, or efficient sporulation (25, 28). It also did not affect transcription of the SW1/SNF-dependent SUC2 gene or the repression of the silent a1/a2 genes, or activation at a distance of a GAL1 promoter (28). It remains to be seen whether the Hho1p protein is really a bona fide H1 or a transcription factor evolutionarily related to the globular domain of histone H1, despite assuming some of the functional properties of H1 in an in vitro assay (25). These and other studies suggest strongly that H1 and its variants probably have roles beyond that of simple repression and stabilization of higher-order structure in chromatin. Gene-specific effects of H1 are clear in one well-documented case, namely, the 5 S rRNA genes of Xenopus laevis. Replacement of the unusual embryonic variant B4 with the somatic H1 during embryogenesis has been shown to be causal for the selective repression of the oocyte 5 S, leaving the somatic genes active (29). Gene-specific effects of H1, in addition to a general, default, repressive role, may turn out to be more common than might have been imagined.
References
1. K. van Holde (1988) Chromatin, Springer-Verlag, New York.
2. A. P. Wolffe (1995) Chromatin: Structure and function, 2nd ed., Academic Press, London.
3. J. O. Thomas and R. D. Kornberg (1975) Proc. Natl. Acad. Sci. USA 72, 2626–2630.
4. G. Arents et al. (1991) Proc. Natl. Acad. Sci. USA 88, 10148–10152.
5. A. Klug et al. (1980). Nature 287, 509–516.
6. K. Luger et al. (1997) Nature 389, 251–260.
7. M. Grunstein (1997) Nature 389, 349–352.
8. L. Huang, W. Z. Zhang, and S. Y. Roth (1997) Mol. Cell. Biol. 17, 6555–6562.
9. P. T. Georgel, T. Tsukiyama, and C. Wu (1997) EMBO J. 16, 4717–4726.
10. M. J. Barratt, C. A. Hazzalin, E. Cano, and L. C. Mahadevan (1994) Proc. Natl. Acad. Sci. USA, 91, 4781–4785.
11. D. Poccia (1986) Int. Rev. Cytol. 105, 1–65.
12. K. F. Sullivan, M. Hechenberger, and K. Masri (1994) J. Cell Biol. 127, 581–592.
13. C. Costanzi and J. R. Pehrson (1998) Nature 393, 599–601.
14. E. Mohr, L. Trieschmann, and U. Grossbach (1989) Proc. Natl. Acad Sci USA 86, 9308–9312.
15. S. Khochbin and A. P. Wolffe (1994) Eur. J. Biochem. 225, 501–510.
16. V. Ramakrishnan (1994) Curr. Opin. Struct. Biol. 4, 44–50.
17. F. A. Goytisolo et al. (1996) EMBO J. 15, 3421–3429.
18. Y.-B. Zhou et al. (1998) Nature 395, 402–405.
19. P. Varga-Weisz et al. (1994) Proc. Natl. Acad. Sci. USA 91, 3525–3529.
20. D. J. Clark, C. S. Hill, S. R. Martin, and J. O. Thomas (1988) EMBO J. 7, 69–75.
21. R. W. Lennox and L. H. Cohen (1988) In Chromosomes and Chromatin, Vol. 1 (K. W. Adolph, ed.) CRC Press, Boca Raton, FL. pp. 33–56.
22. S. Y. Roth and C. D. Allis (1992) Trends Biochem. Sci. 17, 93–98.
23. X. T. Shen and M. A. Gorovsky (1996) Cell 86, 475–483.
24. S. C. Ushinsky et al. (1997) Yeast 13, 151–161.
25. H. G. Patterton et al. (1998) J. Biol. Chem. 273, 7268–7276.
26. A. M. Sirotkin et al. (1995) Proc. Natl. Acad. Sci. USA 92, 6434–6438.
27. M. Dasso, S. Dimitrov, and A. P. Wolffe (1994) Proc. Natl. Acad Sci. USA 91, 12477–12481.
28. D. Escher and W. Schaffner (1997) Mol. Gen. Genet. 256, 456–461.
29. P. Bouvet, S. Dimitrov, and A. P. Wolffe (1994) Genes Dev. 8, 1147–1159.



|
|
|
|
علامات بسيطة في جسدك قد تنذر بمرض "قاتل"
|
|
|
|
|
|
|
أول صور ثلاثية الأبعاد للغدة الزعترية البشرية
|
|
|
|
|
|
|
مكتبة أمّ البنين النسويّة تصدر العدد 212 من مجلّة رياض الزهراء (عليها السلام)
|
|
|