


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date:
Date: 14-4-2021
Date: 6-6-2021
|
Polycomb and Trithorax Are Antagonistic Repressors and Activators
KEY CONCEPTS
- Polycomb group proteins (Pc-G) perpetuate a state of repression through cell divisions.
- A Polycomb response element (PRE) is a DNA sequence that is required for the action of Pc-G.
- The PRE provides a nucleation center from which Pc-G proteins propagate an inactive structure in order to form an epigenetic memory mediated by PREs.
- Trithorax group proteins (TrxG) antagonize the actions of the Pc-G.
- Pc-G and TrxG can bind to the same PRE with opposing effects.
Regions of constitutive heterochromatin, such as at telomeres and centromeres, provide one example of the specific repression of chromatin. Another is provided by the genetics of homeotic genes (which affect the identity of body segments) in Drosophila, which has led to the identification of a protein complex that may maintain certain genes in a repressed state. Polycomb (Pc) mutants show transformations of cell type that are equivalent to gain-of-function mutations in the genes Antennapedia (Antp) or Ultrabithorax, because these genes are expressed in tissues in which they are usually repressed. This implicates Pc in negatively regulating transcription. Furthermore, Pc is the prototype for a class of about 15 loci called the Pc-group (Pc-G); mutations in these genes generally have the same result of derepressing homeotic genes, which suggests the possibility that the group of proteins has some common regulatory role.
The Pc proteins function in large complexes. PRC1 (Polycomb repressive complex 1) contains Pc itself, several other Pc-G proteins, and five general transcription factors. The Esc-E(z) complex contains Esc (extra sex combs), E(z) (enhancer of zeste), other Pc-G proteins, a histone-binding protein, and a histone deacetylase. Pc itself has a chromodomain that binds to methylated H3, and E(z) is a methyltransferase that trimethylates histone H3K27. These properties directly support the connection between chromatin remodeling and repression that was initially suggested by the properties of brahma, a fly counterpart to SWI2.
The brahma gene encodes a component of the SWI/SNF remodeling complex (see the Eukaryotic Transcription Regulation chapter), and loss of brahma function suppresses mutations in Polycomb.
Consistent with the pleiotropy of Pc mutations, Pc is a nuclear protein that can be visualized at approximately 80 sites on polytene chromosomes. These sites include the Antp gene. Another member of the Pc-G, polyhomeotic, is visualized at a set of polytene chromosome bands that are identical to those bound by Pc. The two proteins coimmunoprecipitate in a complex of approximately 2.5 × 106 Da that contains 10 to 15 polypeptides. The relationship between these proteins and the products of the 28 or so Pc-G genes remains to be established. One possibility is that some of these gene products form a general repressive complex, and then some of the other proteins associate with it to determine its specificity.
The Pc-G proteins are not conventional repressors. They are not responsible for determining the initial pattern of expression of the genes on which they act. In the absence of Pc-G proteins, these genes are initially repressed as usual, but later in development the repression is lost without Pc-G group functions. This suggests that the Pc-G proteins in some way recognize the state of repression when it is established, and they then act to perpetuate it through cell division of the daughter cells. FIGURE 1 shows a model in which Pc-G proteins bind in conjunction with a repressor, but the Pc-G proteins remain bound after the repressor is no longer available. This is necessary to maintain repression; otherwise, the gene becomes activated if Pc-G proteins are absent.
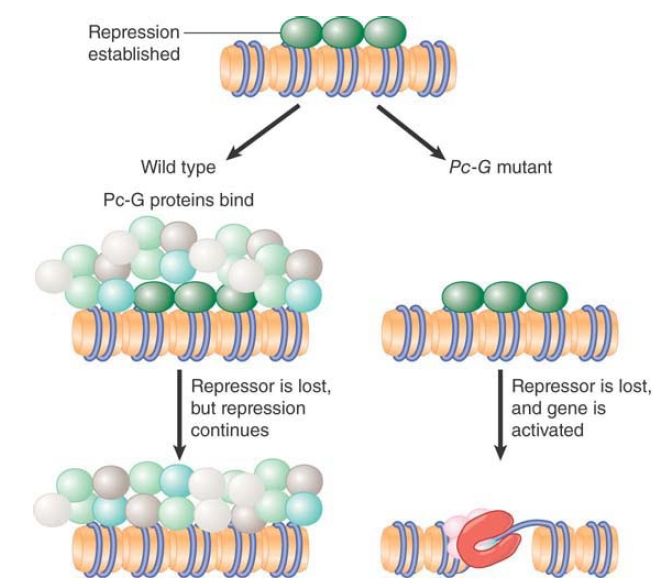
FIGURE 1. Pc-G proteins do not initiate repression, but they are responsible for maintaining it. A Polycomb response element (PRE) is a region of DNA that is sufficient to enable the response to the Pc-G genes. It can be defined operationally by the property that it maintains repression in its vicinity throughout development. The assay for a PRE is to insert it close to a reporter gene that is controlled by an enhancer that is repressed in early development, and then to determine whether the reporter becomes expressed subsequently in the descendants. An effective PRE will prevent such re-expression. The PRE is a complex sequence that measures about 10 kb.
Several proteins with DNA-binding activity for sites within the PRE, including Pho, Pho1, and GAGA factor (GAF), have been identified, but there could be others. When a locus is repressed by Pc-G, however, the Pc-G proteins occupy a much larger length of DNA than the PRE itself. Pc is found locally over a few kilobases of DNA surrounding a PRE. This suggests that the PRE may provide a nucleation center from which a structural state depending on Pc-G proteins may propagate. This model is supported by the observation of effects related to PEV ; that is, a
gene near a locus whose repression is maintained by Pc-G may become heritably inactivated in some cells but not others. In one typical situation, crosslinking experiments in vivo show that Pc protein is found over large regions of the bithorax complex locus that are inactive, but the protein is excluded from regions that contain active genes. The idea that this could be due to cooperative interactions within a multimeric complex is supported by the existence of mutations in Pc that change its nuclear distribution and abolish the ability of other Pc-G members to localize in the nucleus.
The role of Pc-G proteins in maintaining, as opposed to establishing, repression must mean that the formation of the complex at the PRE also depends on the local state of gene expression. The effects of Pc-G proteins are vast in that hundreds of potential Pc-G targets in plants, insects, and mammals have been identified. A working model for Pc-G binding at a PRE is suggested by the properties of the individual proteins. First, Pho and Pho1 bind to specific sequences within the PRE. Esc-E(z) is recruited to Pho/Pho1; it then uses its methyltransferase activity to methylate K27 of histone H3. This creates the binding site for the PRC1, because the chromodomain of Pc binds to the methylated lysine. The dRING component of PRC1 then monoubiquitinates histone H2A on K119, which is linked to chromatin compaction and RNA polymerase II pausing. In addition, long intergenic noncoding RNAs (lincRNAs) play an important role in assembly of Polycomb complexes. For example, the HOTAIR lincRNA acts as a scaffold for assembly of the PRC2 complex . The Polycomb complex induces a more compact structure in chromatin; each PRC1 complex causes about three nucleosomes to become less accessible.
In fact, the chromodomain was first identified as a region of homology between Pc and the protein HP1 found in heterochromatin. Binding of the chromodomain of Pc to K27 on H3 is analogous to HP1’s use of its chromodomain to bind to methylated K9. Variegation is caused by the spreading of inactivity from constitutive heterochromatin, and as a result it is likely that the chromodomain is used by Pc and HP1 in a similar way to induce the formation of heterochromatic or inactive structures. This model implies that similar mechanisms are used to repress individual loci or to create heterochromatin.
In contrast, Trithorax group (TrxG) proteins have the opposite effect of Pc-G proteins: They act to maintain genes in an active state. TrxG proteins are quite diverse; some comprise subunits of chromatin-remodeling enzymes such as SWI/SNF, whereas others also possess important histone-modification activities (such as histone demethylases), which could oppose the activities of Pc-G proteins. The actions of the two groups may share some similarities: Mutations in some loci prevent both Pc-G and TrxG from functioning, suggesting that they could rely on common components. The GAGA factor, which is encoded by the Trithoraxlike gene, has binding sites in the PRE. In fact, the sites where Pc binds to DNA coincide with the sites where GAGA factor binds. What does this mean? GAGA is probably needed for activating factors, including TrxG members, to bind to DNA. Is it also needed for Pc-G proteins to bind and exercise repression? This is not yet clear, but such a model would demand that something other than GAGA determines which of the alternative types of complex subsequently assemble at the site.
The TrxG proteins act by making chromatin continuously accessible to transcription factors. Although Pc-G and TrxG proteins promote opposite outcomes, they bind to the same PREs, which can regulate homeotic gene promoters some distance away from the PRE through looping of DNA.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
المجمع العلمي ينظّم ندوة حوارية حول مفهوم العولمة الرقمية في بابل
|
|
|