


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 27-12-2015
Date: 25-5-2021
Date: 15-11-2020
|
Promoter Activation Involves Multiple Changes to Chromatin
KEY CONCEPTS
- Remodeling complexes can facilitate binding of acetyltransferase complexes, and vice versa.
- Histone methylation can also recruit chromatin-modifying complexes.
-Different modifications and complexes facilitate transcription elongation.
FIGURE 1. summarizes three common differences between active chromatin and inactive chromatin:
- Active chromatin is acetylated on the tails of histones H3 and H4.
- Inactive chromatin is methylated on specific lysines (such as K9) of histone H3.
- Inactive chromatin is methylated on cytosines of CpG doublets.
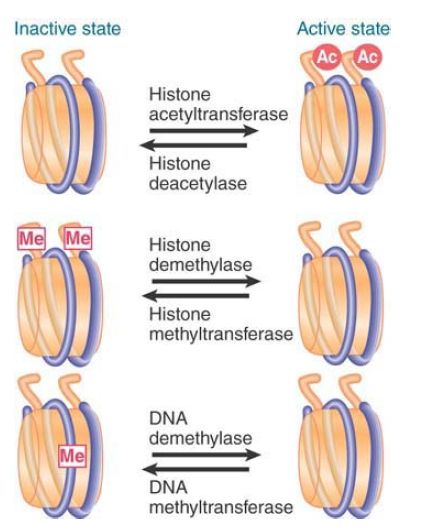
FIGURE 1. Acetylation of histones activates chromatin; methylation of DNA and specific sites on histones inactivates chromatin.
The reverse events occur in the activation of a promoter with the generation of heterochromatin. The actions of the enzymes that modify chromatin ensure that activating events are mutually exclusive with inactivating events. For example, the silencing methylation of H3 at K9 and the activating acetylation of H3 at K9 and K14 are mutually antagonistic.
How are histone-modifying enzymes such as acetyltransferases or deacetylases recruited to their specific targets? As with remodeling complexes, the process is likely to be indirect. A sequence-specific activator (or repressor) may interact with a component of the acetyltransferase (or deacetylase) complex to recruit it to a promoter.
Direct interactions also take place between remodeling complexes and histone-modifying complexes. Histone modifications by themselves have little effect on the overall structure or accessibility of chromatin, which instead requires the interactions of chromatin remodelers. Binding by the SWI/SNF remodeling complex may lead, in turn, to binding by the SAGA acetyltransferase complex.
Acetylation of histones can then stabilize the association with the SWI/SNF complex (via its bromodomain), making a mutual reinforcement of the changes in the components at the promoter. In fact, the Brg1 ATPase subunit of the human SWI/SNF complex requires H4K8 and K12 acetylation for binding to certain targets in vivo. Some remodeling complexes contain between 4 and 10 bromodomains distributed among different subunits, which may confer different binding specificities for specific acetylated targets.
Histone methylation also results in recruitment of numerous factors that contain methyl-lysine recognition motifs such as chromodomains and plant homeodomain (PHD) fingers. Methylation of histone H3 on K4 recruits the chromodomain-containing remodeler Chd1, which also associates with SAGA. H3K4me also directly recruits another acetyltransferase complex, NuA3, which recognizes H3K4me via a PHD domain in one of its subunits. These are just a few of the interactions that occur during transcription activation, and different genes have different (but often overlapping) complex networks of interactions. A further set of dynamic modifications and interactions serves to facilitate transcriptional elongation and to “reset” the chromatin behind the elongating polymerase.
Many of the events at the promoter can be connected into the series illustrated in FIGURE 2 . The initiating event is the binding of a sequence-specific component, which is either able to find its target DNA sequence in the context of chromatin or to bind to a site in a nucleosome-free region. This activator recruits remodeling and histone-modifying complexes (only HATs are shown for simplicity). Changes occur in nucleosome structure, and the acetylation or other modification of target histones provides a covalent mark that the locus has been activated. Many of these steps are mutually reinforcing. Initiation complex assembly follows (after any other necessary activators bind), and at some point histones are typically displaced.
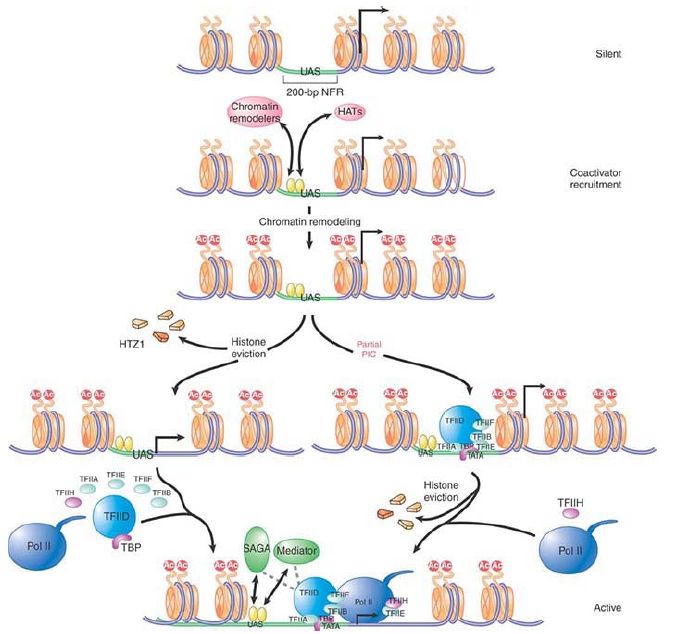
FIGURE 2. Htz1-containing nucleosomes flank a 200-bp NFR on both sides of a promoter. Upon targeting to the upstream activation sequence (UAS), activators recruit various coactivators (such as Swi/Snf or SAGA). This recruitment further increases the binding of activators, particularly for those bound within nucleosomal regions. More important, histones are acetylated at promoter-proximal regions, and these nucleosomes become much more mobile. In one model (left), a combination of acetylation and chromatin remodeling directly results in the loss of Htz1-containing nucleosome, thereby exposing the entire core promoter to the GTFs and Pol II. SAGA and Mediator then facilitate preinitiation complex (PIC) formation through direct interactions. In the other model (right), which represents the remodeled state, partial PICs could be assembled at the core promoter without loss of Htz1. It is the binding of Pol II and TFIIH that leads to the displacement of Htz1-containing nucleosomes and the full assembly of PIC.
Reprinted from Cell, vol. 128, B. Li, M. Carey, and J. L. Workman, The Role of Chromatin
during Transcription, pp. 707–719. Copyright 2007, with permission from Elsevier
[http://www.sciencedirect.com/science/journal/00928674].



|
|
|
|
علامات بسيطة في جسدك قد تنذر بمرض "قاتل"
|
|
|
|
|
|
|
أول صور ثلاثية الأبعاد للغدة الزعترية البشرية
|
|
|
|
|
|
|
مكتبة أمّ البنين النسويّة تصدر العدد 212 من مجلّة رياض الزهراء (عليها السلام)
|
|
|