


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 13-12-2015
Date: 21-4-2016
Date: 5-6-2021
|
Strand-Transfer Proteins Catalyze Single-Strand Assimilation
KEY CONCEPT
- RecA forms filaments with single-stranded or duplex DNA and catalyzes the ability of a single-stranded DNA with a free 3′ end to displace its counterpart in a DNA duplex.
The E. coli protein RecA was the first example of a DNA strandtransfer protein to be discovered. It is the paradigm for a group that includes several other bacterial and archaeal proteins, as well as eukaryotic Rad51 and the meiotic protein Dmc1 . Analysis of yeast rad51
mutants shows that this class of protein plays a central role in recombination. They accumulate DSBs and fail to form normal synaptonemal complexes. This reinforces the idea that exchange of strands between DNA duplexes is involved in formation of the synaptonemal complex and raises the possibility that chromosome synapsis is related to the bacterial strand assimilation reaction.
RecA in bacteria has two quite different types of activity: It can stimulate protease activity in the SOS response , and it can promote base pairing between a single strand of DNA and its complement in a duplex molecule. Both activities are activated by single-stranded DNA in the presence of ATP.
The DNA-handling activity of RecA enables a single strand to displace its homolog in a duplex in a reaction that is called singlestrand assimilation (or single-strand invasion). The displacement reaction can occur between DNA molecules in several configurations and has three general conditions:
- One of the DNA molecules must have a single-stranded region.
- One of the molecules must have a free 3′ end.
- The single-stranded region and the 3′ end must be located within a region that is complementary between the molecules.
The reaction is illustrated in FIGURE 1. When a linear single strand invades a duplex, it displaces the original partner to its complement. The reaction can be followed most easily by making either the donor or recipient a circular molecule. The reaction proceeds 5′→3′ along the strand whose partner is being displaced and replaced; that is, the reaction involves an exchange in which (at least) one of the exchanging strands has a free 3′ end.
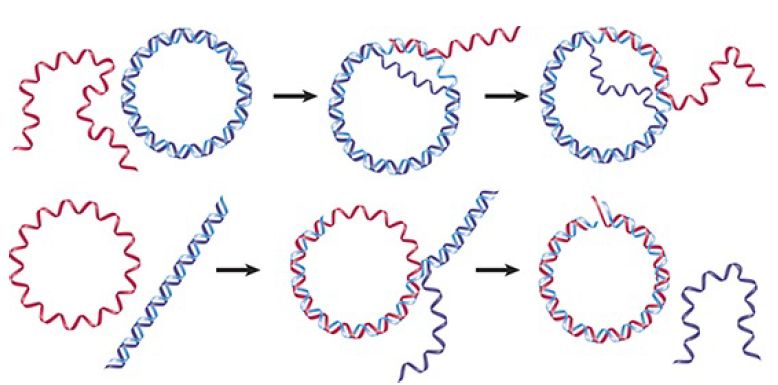
FIGURE 1. RecA promotes the assimilation of invading single strands into duplex DNA as long as one of the reacting strands has a free end.
Single-strand assimilation is potentially related to the initiation of recombination. All models call for an intermediate in which one or both single strands cross over from one duplex to the other . RecA could catalyze this stage of the reaction. In the bacterial context, RecA acts on substrates generated by RecBCD.
RecBCD-mediated unwinding and cleavage can be used to generate ends that initiate the formation of heteroduplex joints. RecA can take the single strand with the 3′ end that is released when RecBCD cuts at chi, and then use it to react with a homologous duplex sequence, thus creating a joint molecule.
All of the bacterial and archaeal proteins in the RecA family can aggregate into long filaments with single-stranded or duplex DNA. Six RecA monomers are bound to DNA per turn of the RecA-DNA filament, which has a helical structure with a deep groove that contains the DNA. The stoichiometry of binding is three nucleotides(or base pairs ) per RecA monomer. The DNA is held in a form that is extended 1.5 times relative to duplex B DNA, making a turn every 18.6 nucleotides (or base pairs). When duplex DNA is bound, it contacts RecA via its minor groove, leaving the major groove accessible for possible reaction with a second DNA molecule.
The interaction between two DNA molecules occurs within these filaments. When a single strand is assimilated into a duplex, the first step is for RecA to bind the single strand into a presynaptic filament. The duplex is then incorporated, probably forming some sort of triple-stranded structure. In this system, synapsis precedes physical exchange of material, because the pairing reaction can take place even in the absence of free ends, when strand exchange is impossible. A free 3′ end is required for strand exchange. The reaction occurs within the filament, and RecA remains bound to the strand that was originally single, so that at the end of the reaction RecA is bound to the duplex molecule.
All of the proteins in this family can promote the basic process of strand exchange without a requirement for energy input. RecA, however, augments this activity by using ATP hydrolysis. Large amounts of ATP are hydrolyzed during the reaction. The ATP may act through an allosteric effect on RecA conformation. When bound to ATP, the DNA-binding site of RecA has a high affinity for DNA; this is needed to bind DNA and for the pairing reaction. Hydrolysis of ATP converts the binding site to low affinity, which is needed to release the heteroduplex DNA.
We can divide the reaction that RecA catalyzes between singlestranded and duplex DNA into three phases:
- A slow presynaptic phase in which RecA polymerizes on singlestranded DNA
- A fast pairing reaction between the single-stranded DNA and its complement in the duplex to produce a heteroduplex joint
- A slow displacement of one strand from the duplex to produce a long region of heteroduplex DNA
The presence of SSB stimulates the reaction by ensuring that the substrate lacks secondary structure. It is not clear yet how SSB and RecA both can act on the same stretch of DNA. Like SSB, RecA is required in stoichiometric amounts, which suggests that its action in strand assimilation involves binding cooperatively to DNA to form a structure related to the filament.
When a single-stranded molecule reacts with a duplex DNA, the duplex molecule becomes unwound in the region of the recombinant joint. The initial region of heteroduplex DNA may not even lie in theconventional double- helical form, but could consist of the two strands associated side by side. A region of this type is called a paranemic joint, as compared with the classical intertwined plectonemic relationship of strands in a double helix, depicted in FIGURE 2.. A paranemic joint is unstable; further progress of the reaction requires its conversion to the double-helical form. Thisreaction is equival ent to removing negative supercoils and may require an enzyme that solves the unwinding/rewinding problem by making transient breaks that allow the strands to rotate about each other.
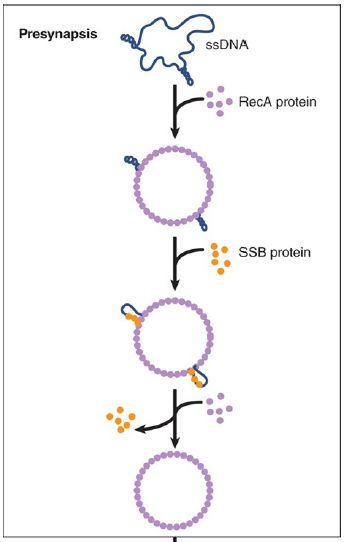

FIGURE 2. Formation of paranemic and plectonemic joints. Once homology is found, side-by-side pairing is formed, called paranemic pairing, which then transitions to plectonemic pairing, where the paired DNA strands are in a double-helix configuration. Note that these pairing stages involve strand invasion and D-loop formation.
Data from P. R. Bianco and S. C. Kowalczykowski. Encyclopedia of Life Sciences. John Wiley & Sons, Ltd., 2005.
All of the reactions we have discussed so far represent only a part of the potential recombination event: the invasion of one duplex by a single strand. Two duplex molecules can interact with each other under the sponsorship of RecA, provided that one of them has a single-stranded region of at least 50 bases. The single-stranded region can take the form of a tail on a linear molecule or of a gap in a circular molecule.
The reaction between a partially duplex molecule and an entirely duplex molecule leads to the exchange of strands. An example is illustrated in FIGURE 3. Assimilation starts at one end of the linear molecule, where the invading single strand displaces its homolog in the duplex in the customary way. When the reaction reaches the region that is duplex in both molecules, though, the invading strand unpairs from its partner, which then pairs with the other displaced strand.
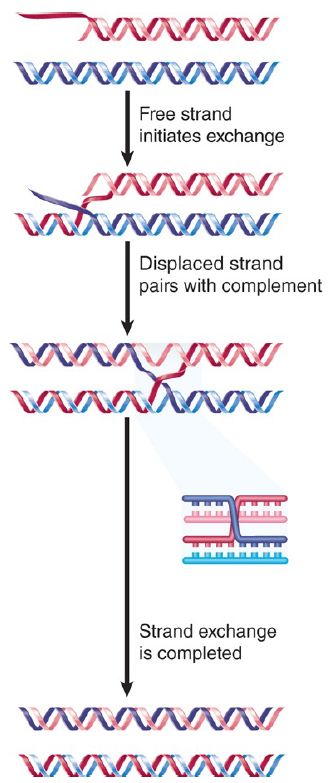
FIGURE 3. RecA-mediated strand exchange between partially duplex and entirely duplex DNA generates a joint molecule with the same structure as a recombination intermediate. At this stage, the molecule has a structure indistinguishable from the recombinant joint . The reaction sponsored in vitro by RecA can generate Holliday junctions, which suggests that the enzyme can mediate reciprocal strand transfer. Less is known about the geometry of the four-strand intermediates bound byRecA, but presumabl y two duplex molecules can lie side by side in a way consistent with the requirements of the exchange reaction.
The biochemical reactions characterized in vitro leave open many possibilities for the functions of strand-transfer proteins in vivo. Their involvement is triggered by the availability of a singlestranded 3′ end. In bacteria, this is most likely generated when RecBCD processes a DSB to generate a single-stranded end. One of the main circumstances in which this is invoked may be when a replication fork stalls at a site of DNA damage . The introduction of DNA during conjugation, when RecA is required for recombination with the host chromosome, is more closely related to conventional recombination. In yeast, DSBs may be generated by DNA damage or as part of the normal process of recombination. In either case, processing of the break to generate a 3′–single-stranded end isfollowed by l oading the single strand into a filament with Rad51, followed by a search for matching duplex sequences. This can be used in both repair and recombination reactions.



|
|
|
|
علامات بسيطة في جسدك قد تنذر بمرض "قاتل"
|
|
|
|
|
|
|
أول صور ثلاثية الأبعاد للغدة الزعترية البشرية
|
|
|
|
|
|
|
مكتبة أمّ البنين النسويّة تصدر العدد 212 من مجلّة رياض الزهراء (عليها السلام)
|
|
|