


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 22-4-2021
Date: 1-5-2016
Date: 22-3-2021
|
min and noc/slm Genes Regulate the Location of the Septum
KEY CONCEPTS
-The location of the septum is controlled by minC, -D, and -E, and by noc/slmA.
-The number and location of septa are determined by the ratio of MinE/MinCD.
-Dynamic movement of the Min proteins in the cell sets up a pattern in which inhibition of Z-ring assembly is highest at the poles and lowest at mid-cell.
-SlmA/Noc proteins prevent septation from occurring in the space occupied by the bacterial chromosome.
Clues to the localization of the septum were first provided by minicell mutants. The original minicell mutation lies in the locus minB. Deletion of minB generates minicells by allowing septation to occur near the poles instead of at mid-cell, and therefore the role of the wild-type minB locus is to suppress septation at the poles.
The minB locus consists of three genes, minC, -D, and -E. The products of minC and minD form a division inhibitor. MinD is required to activate MinC, which prevents FtsZ from polymerizing into the Z-ring.
Expression of MinCD in the absence of MinE, or overexpression even in the presence of MinE, causes a generalized inhibition of division. The resulting cells grow as long filaments without septa.
Expression of MinE at levels comparable to MinCD confines the inhibition to the polar regions, thus restoring normal growth. The determinant of septation at the proper (mid-cell) site is, therefore, the ratio of MinCD to MinE.
The localization activities of the Min system are due to a remarkable dynamic behavior of MinD and MinE, which is illustrated in FIGURE 1. MinD, an ATPase, oscillates from one end of the cell to the other on a rapid time scale. MinD-ATP binds to and accumulates at the bacterial lipid membrane at one pole of the cell, is released, and then rebinds to the opposite pole. The periodicity of this process takes about 30 seconds, so that multiple oscillations occur within one bacterial cell generation. MinC, which cannot move on its own, oscillates as a passenger protein bound to MinD. MinE forms a ring around the cell at the edge of the zone of MinD. The MinE ring moves toward MinD at the poles and is necessary for ATP hydrolysis and the release of MinD from the membrane. The MinE ring then disassembles and reforms at the edge of the MinD zone that forms at the opposite pole. MinD and MinE are each required for the dynamics of the other. The consequence of this dynamic behavior is that the concentration of the MinC inhibitor is lowest at mid-cell and highest at the poles, which directs FtsZ assembly at mid-cell and inhibits its assembly at the poles.
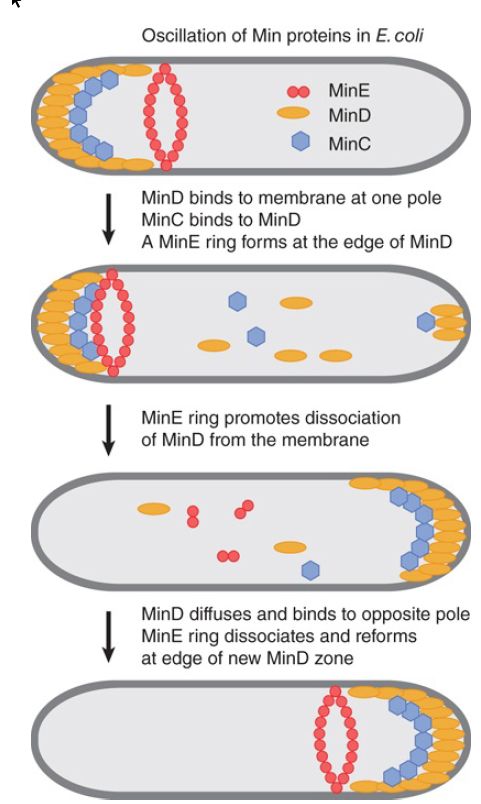
FIGURE 9.8 MinCD is a division inhibitor whose action is confined to the polar sites by MinE.
Another process, called nucleoid occlusion, prevents Z-ring formation over the bacterial chromosome and thus prevents the septum from bisecting an individual chromosome at cell division. A protein called SlmA, which interacts with FtsZ, is necessary for nucleoid occlusion in E. coli. SlmA binds specifically to at least 24 sites on the bacterial chromosome. DNA binding activates SlmA to antagonize the polymerization of FtsZ, which prevents septum formation in this region of the cell. In Bacillus subtilis, a DNAbinding protein called Noc performs a similar nucleoid occlusion role, but by a different mechanism. Noc interacts directly with the membrane, rather than with FtsZ, and this interaction interferes with the assembly of the cell division machinery. The bacterial nucleoid takes up a large volume of the cell, and as a result this process restricts Z-ring assembly to the limited nucleoid-free spaces at the poles and mid-cell. The combination of nucleoid occlusion and the Min system promotes the Z-rings to form, and thus cell division to occur, at mid-cell.



|
|
|
|
كل ما تود معرفته عن أهم فيتامين لسلامة الدماغ والأعصاب
|
|
|
|
|
|
|
ماذا سيحصل للأرض إذا تغير شكل نواتها؟
|
|
|
|
|
|
|
جامعة الكفيل تناقش تحضيراتها لإطلاق مؤتمرها العلمي الدولي السادس
|
|
|