


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 21-3-2021
Date: 28-4-2016
Date: 4-6-2021
|
Introduction to the Clusters and Repeats
A set of genes descended by duplication and variation from a single ancestral gene is called a gene family. Its members can be clustered together or dispersed on different chromosomes (or a combination of both). Genome analysis to identify paralogous sequences shows that many genes belong to families; the 20,000 or so genes identified in the human genome fall into about 15,000 families, so the average gene has about 2 relatives in the genome.
Gene families vary enormously in the degree of relatedness among members, from those consisting of multiple identical members to those for which the relationship is quite distant. Genes are usually related only by their exons, with introns having diverged . Genes can also be related by only some of their exons, whereas others are unique.
Some members of the gene family can evolve to become pseudogenes. Pseudogenes (ψ) are defined by their possession of sequences that are related to those of the functional genes but that cannot be transcribed or translated into a functional polypeptide.
Some pseudogenes have the same general structure as functional genes, with sequences corresponding to exons and introns in the usual locations. They might have been rendered inactive by mutations that prevent any or all of the stages of gene expression.
The changes can take the form of abolishing the signals for initiating transcription, preventing splicing at the exon–intron junctions, or prematurely terminating translation. The initial event that allows the formation of related exons or genes is a duplication, when a copy of some sequence is generated within the genome. Tandem duplication (when the duplicates are in adjacent positions) can arise through errors in replication or recombination. Separation of the duplicates can occur by a translocation that transfers material from one chromosome to another. A duplicate at a new location might also be produced directly by a transposition event that is associated with copying a region of DNA from the vicinity of a transposable element.
Duplications of intact genes, collections of exons, or even individual exons can occur. When an intact gene is involved, duplication generates two copies of a gene whose activities are initially
indistinguishable, but then the copies usually diverge as each accumulates different substitutions.
The members of a structural gene family usually have related or even identical functions, although they might be expressed at different times or in different cell types. For example, different human globin proteins are expressed in embryonic and adult red blood cells, whereas different actins are utilized in muscle and nonmuscle cells. When genes have diverged significantly or when only some exons are related, their products can have different functions.
Some gene families consist of identical members. Clustering is a prerequisite for maintaining identity between genes, although clustered genes are not necessarily identical. Gene clusters range from the extreme case in which a duplication has generated two adjacent related genes to cases in which hundreds of identical genes lie in a tandem array. Extensive tandem repetition of a gene can occur when the product is needed in unusually large amounts.
Examples are the genes encoding rRNA or histone proteins. This creates a special situation with regard to the maintenance of identity and the effects of selective pressure.
Gene clusters offer us an opportunity to examine the forces involved in evolution of the genome over regions larger than single genes. Duplicated sequences, especially those that remain in the same vicinity, provide a means for further evolution by recombination. A population evolves by the classical homologous recombination illustrated in FIGURE .1 and FIGURE .2, in which an exact crossing-over occurs . The recombinant chromosomes have the same organization as the parental chromosome; they contain precisely the same loci in the same order but include different combinations of alleles, providing the raw material for natural selection. However, the existence of duplicated sequences allows aberrant events to occur occasionally, which changes the number of copies of genes and not just the combination of alleles.
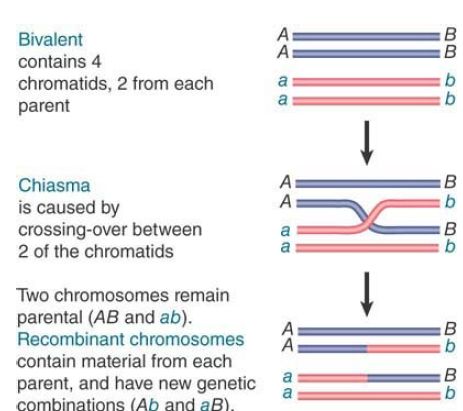
FIGURE .1 Chiasma formation and crossing-over can result in the generation of recombinants.
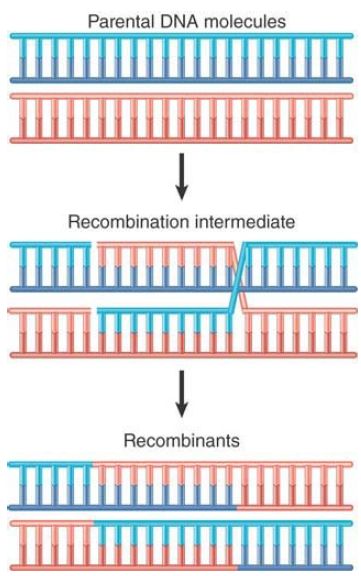
FIGURE .2 Crossing-over and recombination involve pairing between complementary strands of the two parental duplex DNAs.
Unequal crossing over (also known as nonreciprocal recombination) describes a recombination event occurring between two sites that are similar or identical but not precisely aligned. The feature that makes such events possible is the existence of repeated sequences. FIGURE .3 shows that this allows one copy of a repeat in one chromosome to misalign for recombination with a different copy of the repeat in the homologous chromosome instead of with the strictly homologous copy. When recombination occurs, it increases the number of repeats in one chromosome and decreases it in the other. In effect, one recombinant chromosome has a deletion and the other has an insertion. This mechanism is responsible for the evolution of clusters of related sequences. We can trace its operation in expanding or contracting the size of an array in both gene clusters and regions of highly repeated DNA.

FIGURE .3 Unequal crossing-over results from pairing between nonequivalent repeats in regions of DNA consisting of repeating units. Here, the repeating unit is the sequence ABC, and the third repeat of the light-blue chromosome has aligned with the first repeat of the dark-blue chromosome. Throughout the region of pairing, ABC units of one chromosome are aligned with ABC units of the other chromosome. Crossing-over generates chromosomes with 10 and 6 repeats each instead of the 8 repeats of each parent.
The highly repetitive fraction of the genome consists of multiple tandem copies of very short repeating units. These often have unusual properties. One is that they might be identified as a separate peak on a density gradient analysis of DNA); this is the origin of the name satellite DNA because the band containing the repetitive DNA is higher in the gradient than the main band. They often are associated with heterochromatic regions of the chromosomes and in particular with centromeres (which contain the points of attachment for segregation on a mitotic or meiotic spindle). As a result of their repetitive organization, they show some of the same evolutionary patterns as the tandem gene clusters. In addition to the satellite sequences, there are shorter stretches of DNA called minisatellites, tandem repeats in which each repeat is between roughly 10 and 100 base pairs (bp) in length, and they have similar properties. They are useful in showing a high degree of divergence between individual genomes that can be used for mapping or identification purposes.
All of these events that change the constitution of the genome are rare, but they are significant over the course of evolution.



|
|
|
|
لخفض ضغط الدم.. دراسة تحدد "تمارين مهمة"
|
|
|
|
|
|
|
طال انتظارها.. ميزة جديدة من "واتساب" تعزز الخصوصية
|
|
|
|
|
|
|
مشاتل الكفيل تزيّن مجمّع أبي الفضل العبّاس (عليه السلام) بالورد استعدادًا لحفل التخرج المركزي
|
|
|