


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 27-10-2015
Date: 12-11-2015
Date: 11-11-2015
|
CHROMOSOMES
Chromosomes, composed of protein and DNA, are distinct dense bodies found in the nucleus of cells. The chromosomes are named for their ability to take up certain stains (Greek: chromos = coloured, soma = body). Genetic information is contained in the DNA of chromosomes in the form of linear sequences of bases (A, T, C, G). The DNA in an individual chromosome is one, long molecule which is highly coiled and condensed. The total number of bases in all the chromosomes of a human cell is approximately six billion and individual chromosomes range from 50 to 250 million bases. The DNA sequence for a single trait is called a gene. Each chromosome contains a few thousand genes, which range in size from a few thousand bases up to 2 million bases.
The number of chromosomes in human cells is 46, with 22 autosomal pairs (one of each type contributed by the mother and one of each type from the father) and 2 sex chromosomes - Two X chromosomes for females (one from father and one from mother) or an X and a Y chromosome for males (the X from the mother and the Y from the father). The normal chromosomal pattern in the females is 46, XX, and in the males, are 46, XY (Fig. 2). The gametes contain a single set of chromosomes, namely 22 autosomes and one sex chromosome. This single set of chromosomes is called haploid or 1n, in contrast to the chromosome set of a somatic cell, which is diploid, or 2n. At fertilization, each parent contributes a haploid set of chromosomes 1n to the foetus thus restoring the diploid set 2n. Since a male carries two different sex chromosomes X and Y it is clear that if he passes on his X chromosome to the foetus it will be a female foetus (as the contribution from the mother will always be X), and if a father passes on his Y chromosome the foetus will be a male.
Homologous chromosomes have genes at loci in the same sequence though slightly different forms may be present due to polymorphisms on the two different chromosomes. This alternative form of a gene found on the same homologous chromosome is called an allele.
Chromosome morphology
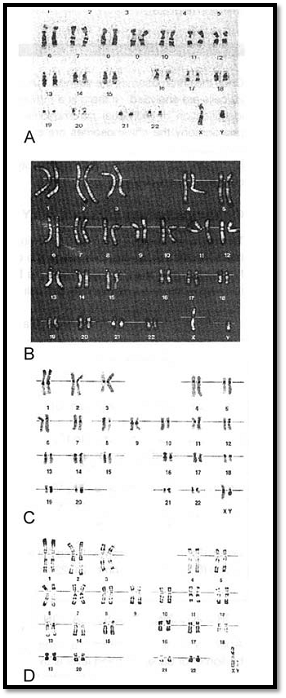
Chromosomes can be visualized by light microscopy. During most of the cell cycle, interphase, the chromosomes are somewhat less condensed and are not visible as individual objects under the light microscope. However, during cell division, mitosis, the chromosomes become highly condensed and are then visible as dark distinct bodies within the nuclei of cells. The chromosomes are most easily seen and identified at the metaphase stage of cell division. The study of chromosomes is called cytogenetics. Various staining techniques have enabled identification of individual chromosomes. An arrangement of chromosomes is called karyotype (Figs 1 and 2).
During metaphase, chromosomes differ from each other in their morphology. Each chromosome is composed of two chromatids joined together at the primary constriction by a centromere. During cell division, the centromere is responsible for cell division. The centromere divides the chromosome into a short and long arm. The part of the chromosome above
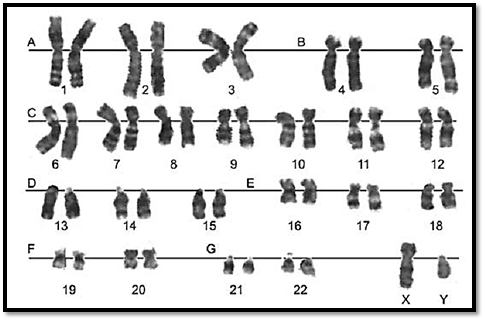
Fig. 1 : GTG banded karyotype from peripheral blood of a normal male showing 22 pairs of autosomes and one pair of sex chromosomes, an X and a Y
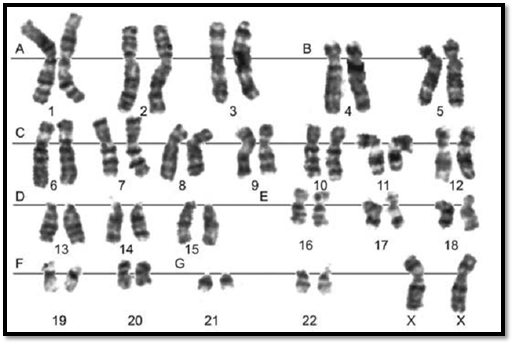
Fig. 2 : GTG banded karyotype from peripheral blood of a normal female showing 22 pairs of autosomes and one pair of sex chromosomes, both chromosome X
the centromere is called short arm (p) and the part below is called long arm (q). The chromosomes are grouped from A to G on the basis of the length of the chromosome and position of the centromere Figure 3. The centromere is either in the middle of a chromosome i.e. metacentric where the short and long arms are equal or is above the centre i.e. sub-metacentric where the p arm is shorter than the q arm, or at the upper end of the chromosome when they are called acrocentric chromosomes. This group has a negligible p arm and a large q arm. The terminal end of a chromosome is called a telomere. Telomeres are specialized structures comprising DNA and protein, which cap the ends of eukaryotic chromosomes.
Besides primary constrictions at the centromere, some of the metaphase chromosomes have secondary constrictions. These secondary constrictions on the acrocentric chromosomes are the site for synthesis of ribosomal material in the interphase nucleus. These regions are termed the Nucleolus Organizer Regions (NORs).
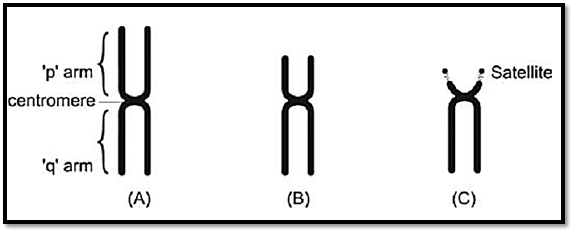
Figs 3A to C: Morphological chromosome classification according to centromere position, (A) Metacentric,(b) Submetacentric, (C) Acrocentric
Methods of chromosome studies
Chromosome Preparation
Chromosomes can be studied from different tissues of the body. The basic principle involved in cytogenetic preparations is the same for all tissues, with slight modifications based on tissue physiology. Constitutional chromosomal patterns are best studied using peripheral blood, which is the most commonly used tissue for cytogenetic investigations. However, skin fibroblasts, and bone marrow are the other types of tissues used. For prenatal diagnosis, chorionic villi, amniotic fluid cells and foetal blood are the tissues that can be used.
Standard Procedures
The basic steps involved in cytogenetic preparation (Fig. 4) include growing the cells in tissue specific media, stimulating undivided cells (T lymphocytes) in blood by a mitogenic agent like phytohemagglutinin for 72 hours, and arresting the spindle formation in cell division by colchicine. In this step, the arrest occurs during metaphase. The chromosomes are in the condensed form in this phase of a cell cycle and the genes located on them cannot be transcribed. This is the most suitable stage for chromosome analysis. The next step is that of harvesting of the sample. In this stage, the cells are given hypotonic treatment so that they swell and chromosomes are released. These are then spread on a slide and can be stained with different staining techniques for visualization and analysis.
In certain acquired haematological malignancies where cells are in a state of spontaneous and continued division, karyotyping from unstimulated blood is also possible. Similarly foetal cord blood or blood from newborns also contain some dividing cells and can be directly karyotyped without stimulation. In prenatal foetal tissues, chorionic villi do not require stimulation as cells of the villi are rapidly dividing and are mitotic. Cells from amniotic fluid also do not require stimulation by mitotic agents but need to be grown for at least ten days before they are ready for harvesting.
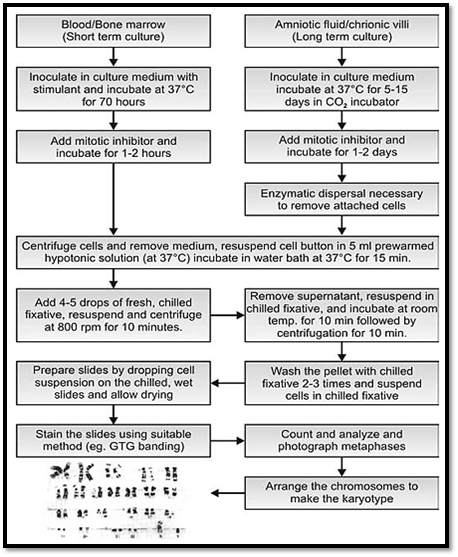
Fig. 4 : Basic steps in cytogenetic preparations
Chromosome Preparation for High Resolution Banding
High Resolution Banding involves banding and staining of chromosomes in prophase or prometaphase. As the chromosomes are more elongated in this phase, the number of bands observable increases to 800 as compared to 400 in the conventional metaphase banding (Fig. 5), thus minor chromosomal aberrations are detectable. In this technique DNA synthesis in the cell is arrested in cell culture to synchronize the cells and release the block. The cultures are then harvested before they condense in late or early prometaphase.
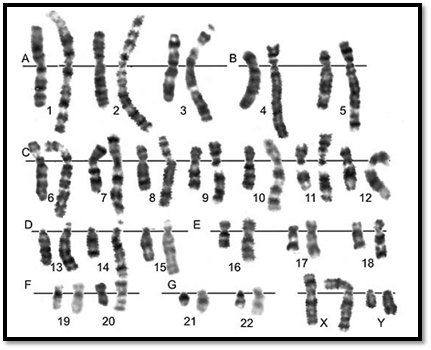
Fig. 5: Comparative karyotypes of routine GTG and high resolution banding
Hereditary Fragile Sites
Several chromosomes have been seen to have fragile sites. They are often harmless on autosomes except for some syndromes and are of significance in chromosomal instability syndromes. The fragile site on the X chromosome at Xq27.3 (Fig. 6) is associated with the fragile X syndrome. This is the most common familial form of mental retardation, and is inherited as an X- linked disorder. This fragile site is rarely expressed in normal culture conditions, and it is expressed by either cultivating cells in folate-deficient medium, or treating the cells with thymidylate synthetase inhibitors such as fluorodeoxyuridine (FudR) in culture. The molecular basis of fragile-X syndrome is now known to be a triplet repeat expansion. DNA analysis of the size of the triplet repeat is now a more widely used method for diagnosis.
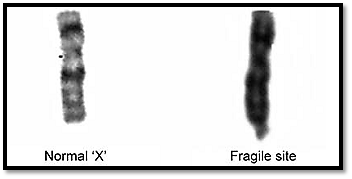
Fig. 6: Fragile site on the X chromosome at Xq27.3
Advantages of using peripheral blood for cytogenetic studies
1. It represents the general constitutional chromosomal pattern of an individual. In rare situations, tissue mosaicism can be suspected if, with a classical clinical picture of a specific syndrome, a normal karyotype is seen. In such cases study from other tissues like skin fibroblasts can help to achieve a diagnosis.
2. Peripheral blood is obtained by simple, minimally invasive and safe technique of blood collection.
3. The culture period is short and gives a good yield.
4. In case of failure in culture growth, re-culturing is possible from the original sample after 4-5 days.
5. Cultures can grow even 48 hours postmortem.
6. A culture is possible in samples mailed by post.
Chromosome staining
The most commonly used stains are Giemsa and Quinacrine. Giemsa is of two types:
1. Conventional Giemsa Stain,
2. Giemsa Trypsin Banding (GTG). The other banding techniques used are R-banding, C-banding and NOR- banding.
Conventional Giemsa Staining
Conventional Giemsa stain is of great value in studying chromosomal morphology (Fig. 7) which includes visualization of satellites, fragile sites, breaks and gaps and to study quadrilateral arrangements as seen in Bloom’s Syndrome. The non-banded spreads are easier to count, and grouping is possible although individual chromosomes cannot be identified.
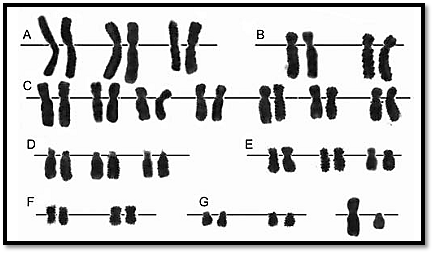
Fig. 7 : Conventional Giemsa staining
Differential Staining Techniques
Giemsa Trypsin Banding (GTG)
In this method, trypsin is used to denature the chromosomal proteins, following which the slides are stained with Giemsa (Fig. 8A). Chromosomes stained by this method show definite patterns of light and dark bands.
Quinacrine Banding (QFQ)
Quinacrine banding is used in identification and structural rearrangement of Y chromosome (Fig. 8B), especially in ambiguous genitalia and in rapid analysis of chromosomal markers in haematological cancers and tumours. Quinacrine heteromorphism can sometimes be useful in identification of maternal v/s foetal cells, donor v/s recipient cells and inherited chromosomal variants.
R Banding
In this method chromosomes receive pre-treatment with heat. The light and dark bands thus produced show a reverse pattern of Giemsa and quinacrine banding (Fig. 8D).
Selective Staining Techniques
C Banding
This method allows selective staining of the constitutive heterochromatin. C-banding is done either by using alkali such as NaOH or Ba(OH) 2. Heteromorphisms in the C-bands are familial, and may be used as markers for certain cases. Unusual morphology in the heteromorphic C-bands, and translocations with a break point in C-banding regions can be identified by this method (Fig. 8C).
NOR Banding
NOR or nucleolar organizing regions are specific chromosomal regions that form and maintain the nucleoli in interphase nuclei. They consist of genes for the larger fraction (28S) of ribosomal RNA. These regions can be stained differentially in metaphase with Giemsa (N-banding) or by silver nitrate (Ag-NOR banding). The N-banding procedure reveals both inactive as well as active NORs, while the Ag-NOR reveals only active NORs. The pattern observed in Ag-NOR banding is consistent for an individual. They can be used in combination with Q-banding to identify paternal origin and the stages of meiotic non-disjunction in trisomies of acrocentrics. Ag-NOR staining has been important in examining the status of the NORs in determining the break points in Robertsonian as well as reciprocal translocations. Silver impregnation can be used to observe changes in activity of NORs in meiosis as well as in malignant cells.
Figs 8A to D: Karyotypes using. (A) G-banding, (B) Q banding, (C) C-banding (D) R-banding
Chromosome classification and analysis
Once the chromosomes are stained, they are ready for analysis. Numerical analysis is done first. A minimum of 20 cells are analyzed. If there is a mosaic cell line, an additional 10,30 or 50 cells are analyzed. Initially, the identification of individual chromosome is done under a microscope. Three fields are then chosen for photography the chromosomes are cut and pasted. This is called a karyotype. In the construction of the karyotype, the arrangement of the autosomes is done in decreasing order of length. The sex chromosomes X and Y are arranged at the end. The karyotyping by conventional photography is now slowly being replaced by computer image processing (Fig. 8).
Chromosome classification by conventional giemsa stain
The earlier methods of chromosome staining allowed identification of chromosomes into seven groups on the basis of their length and position of the centromere (Fig. 1). The autosomes are arranged first in order followed by the sex chromosomes. The following table gives the method of classification of chromosomes stained by conventional solid Giemsa staining.
Chromosome classification by conventional giemsa trypsin banding
A band is defined as that part of a chromosome, which is clearly distinguishable from its adjacent segment. It can be lighter or darker. A band level is the total number of bands countable in a haploid state of chromosomes including sex chromosomes (Table 1). In order to attain a high band level that can detect minor chromosomal defects, long chromosomes are required. This is achieved by studying pro-metaphase
Table 1: Classification of unbanded chromosomes
chromosomes. A technique called high resolution banding is achieved by synchronization of culture followed by a short period of spindle blocking with colchicine, giving a high yield of prometaphase and prophase chromosomes.
Landmark bands, which were first demonstrated in a metaphase spread, were of a 250-band level. In earlier stages of metaphase, 400 band levels can be identified. In a routine cytogenetic laboratory, this band level is sufficient for peripheral blood methods and amniotic cell cultures.
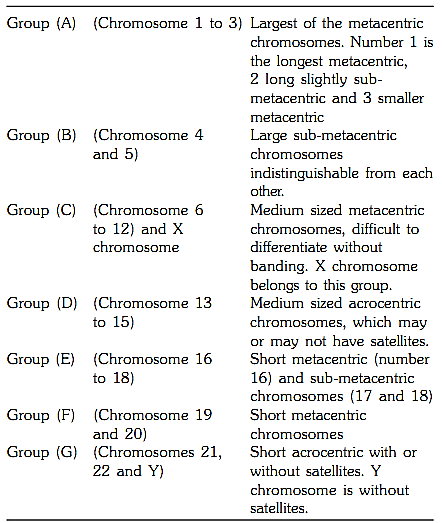
In order to recognize small rearrangements 550 band levels is recommended (prophase or prometaphase stages). 850 band levels can be achieved in longer prophase and prometaphase chromosomes but is not required routinely. ISCN 1995 gives G-banded chromosomes arranged in increasing order of resolution from approximately 500 to 900 band stages (Fig. 9).
Fig.9 : G banded chromosomes arranged in increasing order from 500 to 900 bands. ISCN 1995 classification
Molecular cytogenetics
Fluorescent In Situ Hybridization (FISH)
Conventional staining techniques have limitations in individual assessment of very minute chromosomal structural rearrangements or assessment of sub-microscopic deletions. Fluorescent in situ hybridisation (FISH) is one of the techniques in the field of cytogenetics since the first discovery of chromosomal banding. Denaturation of DNA sequences of metaphase chromosomes, hybridisation of DNA and RNA probes and identification of the target sequence of a chromosome with a probe is the principle of FISH. A fluorochrome tagged receptor molecule binds to DNA probes. A number of fluorochromes are used for this purpose. With the use of fluorescent microscope and special filters, signals are visualized.
In the technique of FISH, nucleic acid sequences of chromosomes (i.e. highly repeated satellite DNA / heterogeneous DNA sequences / specific gene loci sequence) are used as markers and hybridised to chromosomes in a metaphase spread. The technique allows one to detect chromosomal anomalies by specific probes that can be used in prenatal, postnatal or for preimplantation genetic diagnosis.
Diagnostic Applications of FISH
-Identification of specific chromosomes in interphase cells or metaphase spread.
-Identification of individual chromosomes and structural defects, especially microdeletions.
-Gene mapping.
-Identification of species-specific chromosomes by marker probes in hybrids.
-Assessment of radiation effects or damage on individual chromosomes or in metaphase spreads.
-Evaluation of chemical mutagenic effects in individual chromosomes or in metaphase spreads.
Metaphase spread slides are heated with chemicals to break nuclei and remove proteins from chromosomes in order to make open DNA molecules, which will hybridise with DNA probes. After hybridisation, marked probes are removed with a series of washes. Probes are now commercially available for all whole chromosomes, satellite DNAs and many specific loci involved in disease. These and other components can be supplied as kits. Fluorescent microscopes and a special set of filters are required to visualize the FISH results.
PROBES USED IN FISH ANALYSIS
Probes
DNA probes used for the FISH technique are direct-labelled probe and indirect labelled probe. A direct-labelled probe is pre-labelled with the fluorochrome. This probe attaches to the target of interest and allows a fluorescent signal to be bound to the target in the hybridisation stage. An indirect DNA probe is pre-labelled with a hapten. Once this is hybridized to the target sequence fluorochrome labelled antibodies to the hapten are used for probe detection. For this purpose digoxigenin or biotin / streptavidin conjugate is used. The length of the DNA probes used for FISH varies in the range of 20-22 nucleotides to 1Mbp. For detection of short ranges (20-25 nucleotides) synthetic oligomers are used while for tandemly repeated DNA sequences (1 mb) yeast artificial chromosome (YAC) clones are used. Other types of probes used include pools of cosmid contigs, P1 and P1 Derived Artificial Chromosomes (PACs), and Bacterial Artificial Chromosomes (BACs).
Chromosome Enumeration Probes (CEP)
CEP probes are made from chromosome specific sequences from highly repeated human satellite DNA sequences (Fig. 9). The metaphase target chromosome shows a compact, fluorescent spot where the CEP probe hybridises. These probes are useful in determining chromosome specific ploidy in preimplantation, prenatal, postnatal and haematological samples in cultured or uncultured specimens. They are also useful in detecting chromosome specific ploidy in tumour cells, especially in breast and myeloid cancers.
Whole Chromosome Paint Probes (WCP)
DNA probes, which are homologous to DNA sequences of the entire length of an individual chromosome, are called WCP probes. Any one WCP probe is a cocktail of probes for specific DNA sequences of a particular chromosome. These probes are made from chromosome specific recombinant DNA libraries obtained from flow sorted or microdissected, individual chromosomes. The target chromosome looks “painted” by WCP probes (Fig. 10). This helps to recognize an individual chromosome (marker) and identifies translocations, deletions and rearrangements of individual chromosomes in a metaphase spread. WCP probes may be used to detect chromosomal change due to chemical mutagens or radiation damages. It also helps in rapid identification of individual chromosomes in somatic cell hybrids.
Locus Specific Probes (LSI)
The DNA sequences of homologous chromosomes and positions of specific human gene loci can be identified by LSI probes (Fig. 11). The LSI probes are used in tumour, prenatal and postnatal samples.
Multiple Probe Co-hybridisation
Probes labelled with different colour fluoro probes can be mixed together and applied in a single hybridisation to allow simultaneous visualization of two different target sequences on the same nucleus or metaphase spread.
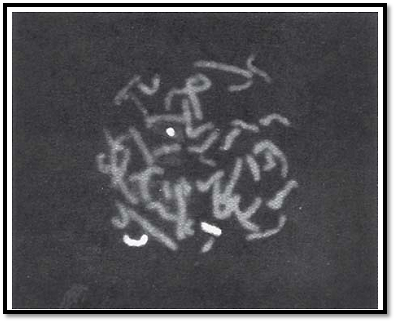
Fig. 10: Fish using whole chromosome paint probes (WCP) showing an 18;21 translocation by dual color chromosome specific probes for chromosome 18 (red) and chromosome 21(green) (For color version see Plate 1)
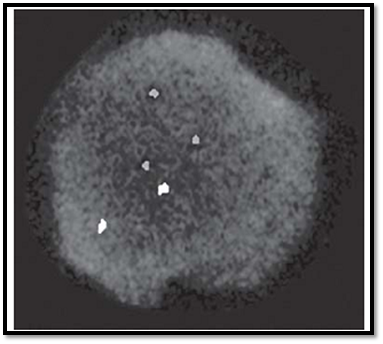
Fig. 11: Interphase FISH using locus specific identifier probes(LSI) Nuclear in situ hybridisation (NUCISH) performed with locus specific probes to detect chromosome 13(green), and chromosome 21(red). Three red signals indicating trisomy
Probes for Telomeric Regions
Telomeric regions of chromosomes are lost due to deletions and unbalanced translocations.
Analysing FISH Results
FISH analysis requires an epiillumination fluorescence microscope.
Application of chromosomal studies
Chromosomal aberrations cannot be corrected, but they are of immense value in diagnosis, prognosis and management of genetic disorders. Some important applications are listed below:
1-To confirm the clinical diagnosis.
2-To identify carrier status of a couple and provide appropriate genetic counselling for prognosis, management and recurrence risk estimation.
3-To plan future prenatal diagnostic tests and consider available reproductive options.
4-In prenatal diagnosis to reassure the couple with normal results or in those with abnormal findings depending on the severity, offer possible options.
5-Karyotyping of products of conception in case of foetal loss may provide a clue to the type of genetic component involved.
6-Cytogenetic studies in malignant tissues especially haematological cancers, help in providing prognosis and assessing the drug response.
7- Chromosomal studies my help in assessment of environmental hazards in Bloom syndrome, Fanconi anaemia and ataxia telangiectasia (explained in the chapter on chromosomal syndromes)
References
Purandarey , H. (2009) . Essentials of Human Genetics. Second Edition. Jaypee Brothers Medical Publishers (P) Ltd.



|
|
|
|
التوتر والسرطان.. علماء يحذرون من "صلة خطيرة"
|
|
|
|
|
|
|
مرآة السيارة: مدى دقة عكسها للصورة الصحيحة
|
|
|
|
|
|
|
نحو شراكة وطنية متكاملة.. الأمين العام للعتبة الحسينية يبحث مع وكيل وزارة الخارجية آفاق التعاون المؤسسي
|
|
|