


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 28-11-2015
Date: 16-12-2015
Date: 17-11-2020
|
Retroelements Fall into Three Classes
KEY CONCEPTS
- LTR retrotransposons mobilize via an RNA that is similar to retroviral RNA but that does not form an infectious particle.
- Although retroelements that lack LTRs, or retroposons, also transpose via reverse transcriptase, they employ a distinct method of integration and are phylogenetically distinct from both retroviruses and LTR retrotransposons.
- Other elements can be found that were generated by an RNA-mediated transposition event, but they do not themselves encode enzymes that can catalyze transposition.
- Retroelements constitute almost half of the human genome.
Retroelements are defined by their use of mechanisms for transposition that involve reverse transcription of RNA into DNA. Three classes of retroelements are distinguished in TABLE .1: LTR retrotransposons, non-LTR retroposons, and the nonautonomous short-interspersed nuclear elements (SINEs).
TABLE .1 Retroelements can be divided into LTR retrotransposons, non-LTR retroposons, and the nonautonomous SINEs.
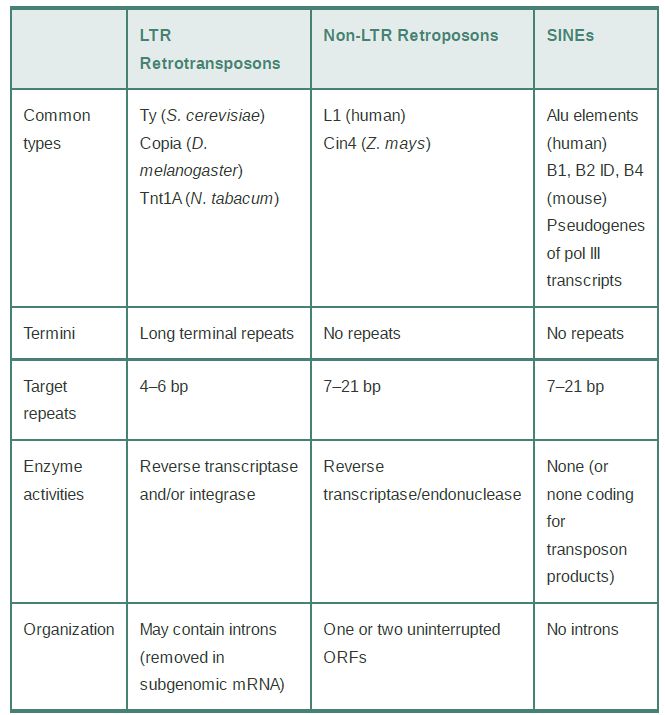
LTR retrotransposons, or simply retrotransposons, have LTRs and encode reverse transcriptase and integrase activities. They reproduce in the same manner as retroviruses but differ from them in not passing through an independent infectious form. They are best characterized in the Ty, copia, and Tos17 elements of yeast, flies, and rice, respectively.
The non-LTR retrotransposons, or retroposons, also have reverse transcriptase activity but constitute a phylogenetically distinct family of elements that employ a distinct transposition mechanism. Unlike retrotransposons and retroviruses, retroposons lack LTRs and use a different mechanism from retroviruses to prime the reverse transcription reaction. They are derived from RNA polymerase II transcripts. Only a few of the elements in a given genome are fully functional and can transpose autonomously; others have mutations, and thus can only transpose as the result of the action of a transacting autonomous element. The most common elements of this class in the human genome are the long-interspersed nuclear elements, or LINEs.
In addition to LTR retrotransposons and non-LTR retroposons, many genomes contain large numbers of sequences whose external and internal features suggest that they originated in RNA
sequences. In these cases, though, we can only speculate about how a DNA copy was generated. We assume that they were targets for a transposition event by an enzyme system coded elsewhere—that is, they are always nonautonomous—and that they originated in cellular transcripts. They do not code for proteins that have transposition functions. The most prominent components of this family are called short-interspersed nuclear elements (SINEs). These elements are derived from RNA polymerase III transcripts, usually 7SL RNAs, 5S rRNAs, and tRNAs. Many of these elements also include portions of a cognate LINE, leading to the hypothesis that SINEs can use the enzymatic machinery of LINEs for replication.
FIGURE 1 shows the organization and sequence relationships of elements that encode reverse transcriptase. Like retroviruses, the LTR retrotransposons can be classified into groups according to the number of independent reading frames for gag, pol, and intand the order of the genes. In spite of these superficial differences of organization, the common features are the presence of LTRs as well as reverse transcriptase and integrase activities. In contrast, non-LTR retroposons such as the mammalian LINEs lack LTRs.
They have two reading frames; one codes for a nucleic acid– binding protein, and the other codes for reverse transcriptase and endonuclease activity.
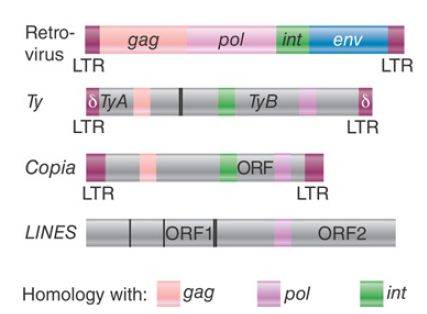
FIGURE 1. Retrotransposons that are closely related to retroviruses have a similar organization, but non-LTR retroposons such as LINEs share only the reverse transcriptase activity and lack LTRs.
LTR-containing elements can vary from integrated retroviruses to retrotransposons that do not have the capacity to generate infectious particles. Yeast and fly genomes have the Ty and copia
elements that cannot generate infectious particles. Mammalian genomes have some endogenous retroviruses that, when active, can generate infectious particles. The mouse genome has several active endogenous retroviruses that are able to generate particles that propagate horizontal infections. By contrast, almost all endogenous retroviruses lost their activity some 50 million years ago in the human lineage, and the genome now has mostly inactive remnants of the endogenous retroviruses.
LINEs and SINEs comprise a major part of the animal genome. They were defined originally by the existence of a large number of relatively short sequences that are related to one another. They are described as interspersed sequences or interspersed repeats because of their common occurrence and widespread distribution.
In many higher eukaryotic genomes, particularly metazoans, LINEs and SINEs can make up half of the total DNA. In contrast, in plant genomes LTR retrotransposons tend to predominate.
FIGURE 2 summarizes the distribution of the different types of transposons that constitute almost half of the human genome. Except for the SINES, which never encode functional proteins, the other types of elements all consist of functional elements and elements that have suffered deletions that eliminated parts of the reading frames that code for the protein(s) needed for transposition. The relative proportions of these types of transposons are generally similar in the mouse genome.
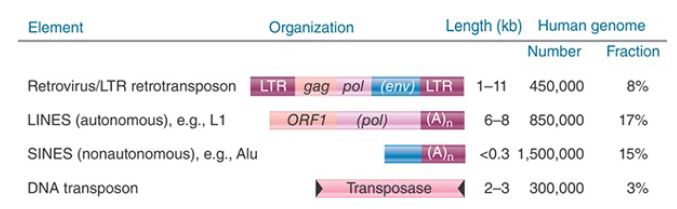
FIGURE 2. Four types of transposable elements constitute almost half of the human genome.
The most common LINE in mammalian genomes is called L1. The typical member is about 6,500 bp long and terminates in a tract rich in adenine. The two open reading frames of a full-length element are called ORF1 and ORF2. The number of full-length elements is usually small (around 50), and the remainder of the copies are truncated. Transcripts can be found. As implied by its presence in repetitive DNA, the LINE family shows sequence variation among individual members. The members of the family within a species, however, are relatively homogeneous compared to the variation shown between species. L1 is the only member of the LINE family that has been active in either the mouse or human lineages. It seems to have remained highly active in the mouse, but has declined in the human lineage.
Only one SINE has been active in the human lineage: the common Alu element. The mouse genome has a counterpart to this element (B1) and also other SINES (B2, ID, B4) that have been active. Human Alu and mouse B1 SINEs are probably derived from the 7SL RNA (see the section later in this chapter titled The Alu Family Has Many Widely Dispersed Members). The other mouse SINEs appear to have originated from reverse transcripts of tRNAs. The transposition of the SINES probably results from their recognition as substrates by an active L1 element.



|
|
|
|
علامات بسيطة في جسدك قد تنذر بمرض "قاتل"
|
|
|
|
|
|
|
أول صور ثلاثية الأبعاد للغدة الزعترية البشرية
|
|
|
|
|
|
|
العتبة الحسينية تطلق فعاليات المخيم القرآني الثالث في جامعة البصرة
|
|
|