


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 10-5-2016
Date: 6-6-2021
Date: 25-4-2016
|
DNA Polymerases Control the Fidelity of Replication
KEY CONCEPTS
-High-fidelity DNA polymerases involved in replication have a precisely constrained active site that favors binding of Watson–Crick base pairs.
-DNA polymerases often have a 3′–5′ exonuclease activity that is used to excise incorrectly paired bases.
-The fidelity of replication is improved by proofreading by a factor of about 100.
The fidelity of replication poses the same sort of problem encountered in considering (for example) the accuracy of translation. It relies on the specificity of base pairing. Yet when we consider the energetics involved in base pairing, we would expect errors to occur with a frequency of approximately 10-2 per base pair replicated. The actual rate in bacteria seems to be approximately 10-8 to 10-10 . This corresponds to about 1 error per genome per 1,000 bacterial replication cycles, or approximately 10-6 per gene per generation.
Researchers can divide the errors that DNA polymerase makes during replication into two classes:
-Substitutions occur when the wrong (improperly paired) nucleotide is incorporated. The error level is determined by the efficiency of proofreading, in which the enzyme scrutinizes the newly formed base pair and removes the nucleotide if it is mispaired.
-Frameshifts occur when an extra nucleotide is inserted or omitted. Fidelity with regard to frameshifts is affected by the processivity of the enzyme: the tendency to remain on a single template rather than to dissociate and reassociate. This is particularly important for the replication of a homopolymeric stretch—for example, a long sequence of dTn :dAn —in which “replication slippage” can change the length of the homopolymeric run. As a general rule, increased processivity reduces the likelihood of such events. In multimeric DNA polymerases, processivity is usually increased by a particular subunit that is not needed for catalytic activity per se.
Bacterial replication enzymes have multiple error reduction systems. The geometry of an A-T base pair is very similar to that of a G-C base pair, as is discussed in the chapter Genes Are DNA and Encode RNAs and Polypeptides. This geometry is used by high-fidelity DNA polymerases as a fidelity mechanism. Only an incoming dNTP that base pairs properly with the template nucleotide fits in the active site, whereas mispairs such as A-C or A-A have the wrong geometry to fit into the active site. On the other hand, low-fidelity DNA polymerases, such as E. coli DNA polymerase IV used for damage bypass replication, have a more open active site that accommodates damaged nucleotides, but also incorrect base pairs. Thus, either the expression or activity of these error-prone DNA polymerases is tightly regulated so that they are only active after DNA damage occurs.
All of the bacterial enzymes possess a 3′–5′ exonucleolytic activity that proceeds in the reverse direction from DNA synthesis. This provides a proofreading function, as illustrated in FIGURE 11.6. In the chain elongation step, a precursor nucleotide enters the position at the end of the growing chain. A bond is formed. The enzyme moves one base pair (bp) farther and then is ready for the next precursor nucleotide to enter. If a mistake has been made, the DNA is structurally warped by the incorporation of the incorrect base that will cause the polymerase to pause or slow down. This will allow the enzyme to back up and remove the incorrect base. In some regions errors occur more frequently than in others; that is, mutation hotspots occur in the DNA. This is caused by the underlying sequence context; some sequences cause the polymerase to move faster or slower, which affects the ability to catch an error.
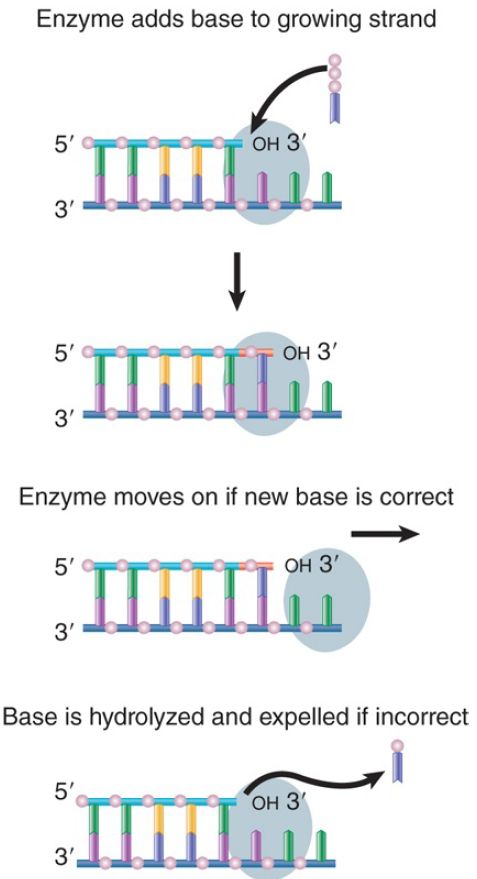
FIGURE 1. DNA polymerases scrutinize the base pair at the end of the growing chain and excise the nucleotide added in the case of a misfit.
As noted in the section DNA Polymerases Are the Enzymes That Make DNA earlier in this chapter, replication enzymes typically are found as multisubunit holoenzyme complexes, whereas repair DNA polymerases are typically found as single subunit enzymes. An advantage to a holoenzyme system is the availability of a specialized subunit responsible for error correction. In E. coli DNA polymerase III, this activity, a 3′ to 5′ exonuclease, resides in a separate subunit, the ε subunit. This subunit gives the replication enzyme a greater fidelity than the repair enzymes.
Different DNA polymerases handle the relationship between the polymerizing and proofreading activities in different ways. In some cases, the activities are part of the same protein subunit, but in others they are contained in different subunits. Each DNA polymerase has a characteristic error rate that is reduced by its proofreading activity. Proofreading typically decreases the error rate in replication from approximately 10-5 to 10-7 /bp replicated.
Systems that recognize errors and correct them following replication then eliminate some of the errors, bringing the overall rate to less than 10-9 /bp replicated .
The replicase activity of DNA polymerase III was originally discovered by a conditional lethal mutation in the dnaE locus, which encodes a 130-kD subunit that possesses the DNA synthetic activity. The 3′–5′ exonucleolytic proofreading activity is found in another subunit, ε, encoded by the dnaQ gene. The basic role of the ε subunit in controlling the fidelity of replication in vivo is demonstrated by the effect of mutations in dnaQ: The frequency with which mutations occur in the bacterial strain is increased by greater than 103 -fold.



|
|
|
|
تفوقت في الاختبار على الجميع.. فاكهة "خارقة" في عالم التغذية
|
|
|
|
|
|
|
أمين عام أوبك: النفط الخام والغاز الطبيعي "هبة من الله"
|
|
|
|
|
|
|
قسم شؤون المعارف ينظم دورة عن آليات عمل الفهارس الفنية للموسوعات والكتب لملاكاته
|
|
|