


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 29-12-2015
Date: 13-12-2015
Date: 7-3-2021
|
Initiation: Creating the Replication Forks at the Origin oriC
KEY CONCEPTS
-Initiation at oriC requires the sequential assembly of a large protein complex on the membrane.
-oriC must be fully methylated.
-DnaA-ATP binds to short repeated sequences and forms an oligomeric complex that melts DNA.
-Six DnaC monomers bind to each hexamer of DnaB, and this complex binds to the origin.
-A hexamer of DnaB forms the replication fork. Gyrase and SSB are also required.
-A short region of A-T-rich DNA is melted.
-DnaG primase is bound to the helicase complex and creates the replication forks.
Initiation of replication of duplex DNA in E. coli at the origin of replication, oriC, requires several successive activities. Some events that are required for initiation occur uniquely at the origin; others recur with the initiation of each Okazaki fragment during the elongation phase :
Protein synthesis is required to synthesize the origin recognition protein, DnaA. This is the E. coli licensing factor that must be made anew for each round of replication. Drugs that block protein synthesis block a new round of replication, but not continuation of replication.
There is a requirement for transcription activation. This is not synthesis of the mRNA for DnaA, but rather either one of two genes that flank oriC must be transcribed. This transcription
near the origin aids DnaA in twisting open the origin.
There must be membrane/cell wall synthesis. Drugs (like penicillin) that inhibit cell wall synthesis block initiation of replication.
Initiation of replication at oriC begins with formation of a complex that ultimately requires six proteins: DnaA, DnaB, DnaC, HU, gyrase, and SSB. Of the six proteins, DnaA draws our attention as the one uniquely involved in the initiation process. DnaB, an ATP hydrolysis-dependent 5′ to 3′ helicase, provides the “engine” of initiation after the origin has been opened (and the DNA is singlestranded) by its ability to further unwind the DNA. These events will only happen if the DNA at the origin is fully methylated on both strands.
DnaA is an ATP-binding protein. The first stage in initiation is binding of the DnaA-ATP protein complex to the fully methylated oriC sequence. This takes place in association with the inner membrane. DnaA is in the active form only when bound to ATP.
DnaA has intrinsic ATPase activity that hydrolyzes ATP to ADP and thus inactivates itself when the initiation stage ends. This ATPase activity is stimulated by membrane phospholipids and singlestranded DNA. Single-stranded DNA forms as soon as the origin is open. This is part of the mechanism used to prevent reinitiation of replication. The origin of the replication region remains attached to the membrane for about one-third of the cell cycle as another part of the mechanism to prevent reinitiation. While sequestered in the membrane, the newly synthesized strand of oriC cannot be methylated and so oriC remains hemimethylated until DnaA is degraded.
Opening oriC involves action at two types of sequence in the origin: 9-bp and 13-bp repeats. Together the 9-bp and 13-bp repeats define the limits of the 245-bp minimal origin, as indicated in FIGURE 1. An origin is activated by the sequence of events summarized in FIGURE 1, in which binding of DnaA-ATP is succeeded by association with the other proteins.
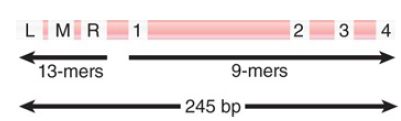
FIGURE 1. The minimal origin is defined by the distance between the outside members of the 13-mer and 9-mer repeats.

FIGURE 2. A two-state assembly model during initiation. DnaAATP monomers in an extended state associate with the high-affinity 13-mer sequences. DnaA-ATP transitions to a compact state as the 9-mer region begins to melt, stabilizing the single-stranded DNA.
Data from: Duderstadt, K. E., et al. 2010. “Origin Remodeling and Opening in Bacteria.” Journal of Biological Chemistry 285:28229–28239, The American Society for Biochemistry and Molecular Biology.
The four 9-bp consensus sequences on the right side of oriC provide the initial binding sites for DnaA-ATP in an extended multimeric state promoted by the accessory protein DiaA, which stimulates cooperative binding of DnaA. DnaA-ATP binds cooperatively to form a helical central core around which oriC DNA is wrapped. DnaA then acts at three A-T–rich 13-bp tandem repeats located on the left side of oriC. In its active form, DnaAATP transitions from the extended state to a compact form, twisting open the DNA strands in an unknown manner to form anopen bubble complex and stabilizing the single-stranded DNA. All three 13-bp repeats must be opened for the reaction to proceed to the next stage. Transcription of either of the two genes flanking oriC provides additional torsional stress to help snap apart the double-stranded DNA.
Altogether, two to four monomers of DnaA-ATP bind at the origin, and after release of DiaA, they recruit two “prepriming” complexes of the DnaB helicase bound to DnaC-ATP, so that there is one DnaB–DnaC-ATP complex for each of the two (bidirectional) replication forks. The function of DnaC is that of a chaperone to repress the helicase activity of DnaB until it is needed. Each DnaB–DnaC complex consists of six DnaC monomers bound to a hexamer of DnaB. Note that the DnaB helicase cannot open double-stranded DNA; it can only unwind DNA that has already been opened, in this case by DnaA. DnaB binding to single-stranded DNA is the signal to hydrolyze ATP and for release of DnaC.
The prepriming complex generates a protein aggregate of 480 kD, which corresponds to a sphere with a radius of 6 nm. The formation of a complex at oriC is detectable in the form of the large protein blob visualized in Figure 2. When replication begins, a replication bubble becomes visible next to the blob. The region of strand separation in the open complex is large enough for both DnaB hexamers to bind, which initiates the two replication forks. As DnaB binds, it displaces DnaA from the 13-bp repeats and extends the length of the open region using its helicase activity. It then uses its helicase activity to extend the region of unwinding. Each DnaB activates a DnaG primase—in one case to initiate the leading strand, and in the other to initiate the first Okazaki fragment of the lagging strand.
Some additional proteins are required to support the unwinding reaction. Gyrase, a type II topoisomerase, provides a swivel that allows one DNA strand to rotate around the other. Without this reaction, unwinding would generate torsional strain (overwinding) in the DNA that would resist unwinding by the helicase. The protein single-strand binding protein (SSB) stabilizes and protects the single-stranded DNA as it is formed and modulates the helicase activity. The length of duplex DNA that usually is unwound to initiate replication is probably less than 60 bp. The protein HU is a general DNA-binding protein in E. coli. Its presence is not absolutely required to initiate replication in vitro, but it stimulates the reaction.
HU has the capacity to bend DNA and is involved in building the structure that leads to formation of the open complex. Input of energy in the form of ATP is required at several stages forthe prepriming reaction, and it is re quired for unwinding DNA. The helicase action of DnaB depends on ATP hydrolysis, and the swivel action of gyrase requires ATP hydrolysis. ATP also is needed for the action of primase and to load the β subunit of Pol III in order to initiate DNA synthesis.
After the prepriming complex is loaded onto the replication forks, the next step is the recruitment of the primase, DnaG, which is then loaded onto the DnaB hexamer. This entails release of DnaC, which allows the DnaB helicase to become active. DnaC hydrolyzes ATP in order to release DnaB. This step marks the transition frominitiation to elongation .



|
|
|
|
علامات بسيطة في جسدك قد تنذر بمرض "قاتل"
|
|
|
|
|
|
|
أول صور ثلاثية الأبعاد للغدة الزعترية البشرية
|
|
|
|
|
|
|
مكتبة أمّ البنين النسويّة تصدر العدد 212 من مجلّة رياض الزهراء (عليها السلام)
|
|
|