


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 29-11-2015
Date: 5-5-2016
Date: 4-11-2020
|
Labelling DNA Gene Probe Molecules
An essential feature of a gene probe is that it can be visualised by some means. In this way, a gene probe that hybridises to a complementary sequence may be detected and identify that desired sequence from a complex mixture. There are two main ways of labelling gene probes; traditionally it has been carried out using radioactive labels, but gaining in popularity are non-radioactive labels. Perhaps the most often used radioactive label is phosphorus-32 (32P), although for certain techniques sulfur-35 (35S) and tritium (3H) are used. These may be detected by the process of autoradiography, where the labelled probe molecule, bound to sample DNA, located for example on a nylon membrane, is placed in contact with an X-ray-sensitive film. Following exposure, the film is developed and fixed just as a black and white negative and reveals the precise location of the labelled probe and therefore the DNA to which it has hybridised.
Non-radioactive labels are increasingly being used to label DNA gene probes. Until recently, radioactive labels were more sensitive than their non-radioactive counterparts. However, recent developments have led to similar sensitivities, which, when combined with their improved safety, have led to their greater acceptance.
The labelling systems are termed either direct or indirect. Direct labelling allows an enzyme reporter such as alkaline phosphatase to be coupled directly to the DNA. Although this may alter the characteristics of the DNA gene probe, they offer the advantage of rapid analysis since no intermediate steps are needed. However, indirect labelling is at present more popular. This relies on the incorporation of a nucleotide which has a label attached. At present, three of the main labels in use are biotin, fluorescein and digoxygenin. These molecules are covalently linked to nucleotides using a carbon spacer arm of 7, 14 or 21 atoms.
Specific binding proteins may then be used as a bridge between the nucleotide and a reporter protein such as an enzyme. For example, biotin incorporated into a DNA fragment is recognised with a very high affinity by the protein streptavidin. This may be either coupled or conjugated to a reporter enzyme molecule such as alkaline phosphatase.
This is able to convert a colourless substrate, p-nitrophenol phosphate (PNPP), into a yellow compound, p-nitrophenol (PNP), and also offers a means of signal amplification. Alternatively labels such as digoxygenin incorporated into DNA sequences may be detected by monoclonal antibodies, again conjugated to reporter molecules including alkaline phosphatase. Thus, rather than the detection system relying on autoradiography, which is necessary for radiolabels, a series of reactions resulting in either a colour or a light or chemiluminescent reaction takes place. This has important practical implications since autoradiography may take 1–3 days, whereas colour and chemiluminescent reactions take minutes.
1- End Labelling of DNA Molecules
The simplest form of labelling DNA is by 5´ or 3´ end labelling; 5´ end labelling involves a phosphate transfer or exchange reaction where the 5´ phosphate of the DNA to be used as the probe is removed and in its place a labelled phosphate, usually 32P, is added. This is usually carried out by using two enzymes; the first, alkaline phosphatase, is used to remove the existing phosphate group from the DNA. Following removal of the released phosphate from the DNA, a second enzyme, polynucleotide kinase, is added, which catalyses the transfer of a phosphate group (32P-labelled) to the 50 end of the DNA. The newly labelled probe is then purified, usually by chromatography through a Sephadex column, and may be used directly (Figure 1).
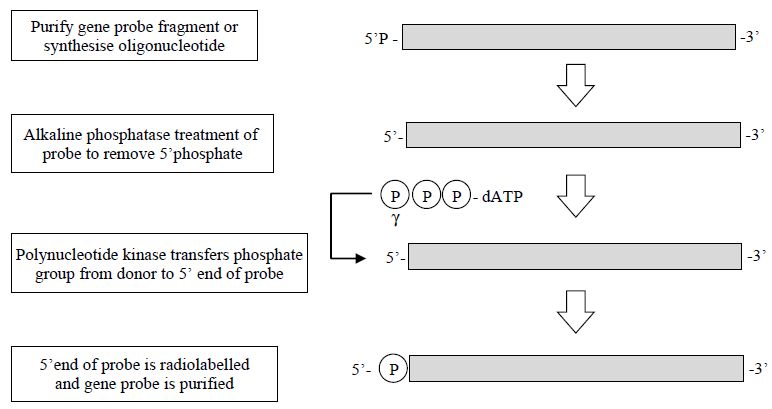
Figure 1. The steps involved in the production of a 5´-labelled gene probe.
Using the other end of the DNA molecule, the 3´ end, is slightly less complex. Here a new dNTP which is labelled (e.g. [32P]adATP or biotinlabelled dNTP) is added to the 3´ end of the DNA by the enzyme terminal transferase. Although this is a simpler reaction, a potential problem exists because a new nucleotide is added to the existing
sequence and so the complete sequence of the DNA is altered, which may affect its hybridisation to its target sequence. End labelling methods also suffer from the fact that only one label is added to the DNA so they are of a lower activity in comparison with methods that incorporate label along the length of the DNA (Figure 2).
2 - Random Primer Labelling
In random primer labelling the DNA to be labelled is first denatured and then placed under renaturing conditions in the presence of a mixture of many different random sequences of hexamers or hexanucleotides.
These hexamers will, by chance, bind to the DNA sample wherever they encounter a complementary sequence and so the DNA will rapidly acquire an approximately random sprinkling of hexanucleotides annealed to it. Each of the hexamers can act as a primer for the synthesis of a fresh strand of DNA catalysed by DNA polymerase since it has an exposed 3´-hydroxyl group. The Klenow fragment of DNA polymerase
is used for random primer labelling because it lacks a 5´–3´ exonuclease

Figure 2. The steps involved in the production of 3´-labelled gene probe.
activity. This is prepared by cleavage of DNA polymerase with subtilisin, giving a large enzyme fragment which has no 5´ to 3´ exonuclease activity, but which still acts as a 5´ to 3´ polymerase. Thus, when the Klenow enzyme is mixed with the annealed DNA sample in the presence of dNTPs, including at least one which is labelled, many short stretches of labelled DNA will be generated (Figure 3). In a similar way to random primer labelling, the polymerase chain reaction may also be used to incorporate radioactive or non-radioactive labels.
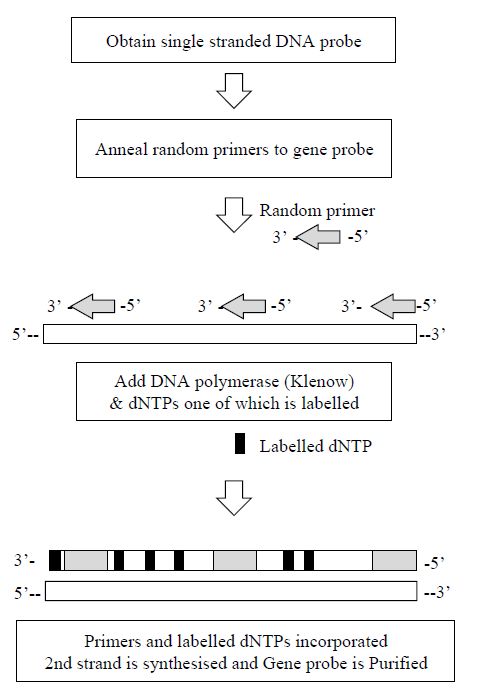
Figure 3. The steps involved in the production of a gene probe produced by the
random hexamer method.
3- Nick Translation
A traditional method of labelling DNA is by the process of nick translation. Low concentrations of DNase I are used to make occasional single-strand nicks in the double-stranded DNA that is to be used as the gene probe. DNA polymerase then fills in the nicks, using an appropriate deoxyribonucleoside triphosphate (dNTP), at the same time making a new nick to the 30 side of the previous one. In this way, the nick is translated along the DNA. If labelled dNTPs are added to the reaction mixture, they will be used to fill in the nicks and so the DNA can be labelled to a very high specific activity (Figure 4).
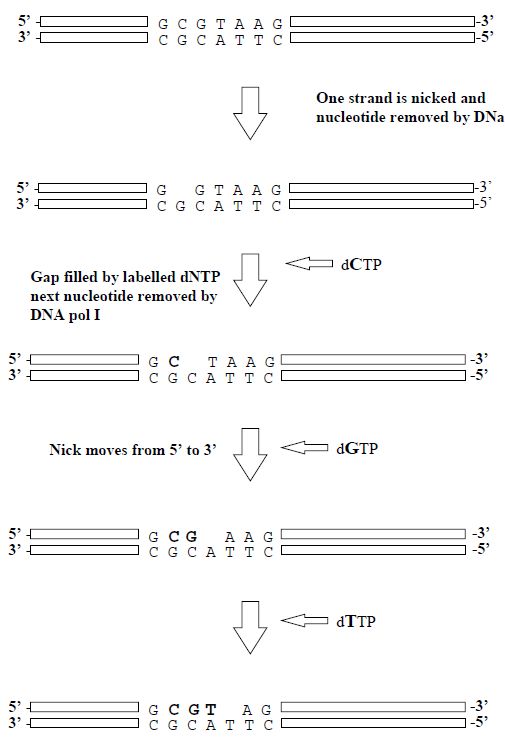
Figure 4. The steps involved in the production of a gene probe by the nick translation method.



|
|
|
|
علامات بسيطة في جسدك قد تنذر بمرض "قاتل"
|
|
|
|
|
|
|
أول صور ثلاثية الأبعاد للغدة الزعترية البشرية
|
|
|
|
|
|
|
مكتبة أمّ البنين النسويّة تصدر العدد 212 من مجلّة رياض الزهراء (عليها السلام)
|
|
|