


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 16-12-2015
Date: 4-4-2021
Date: 2-5-2021
|
Excision Repair
Excision repair (1, 2) is a form of DNA repair involving the removal of a damaged nucleotide from DNA by dual incisions bracketing the damage (3, 4). The multisubunit enzyme system that makes the dual incision is referred to as excision nuclease or excinuclease (3). The damaged nucleotide is released in the form of an oligonucleotide, and the resulting single-stranded gap is filled in by DNA polymerase and sealed by a DNA Ligase. This repair mechanism, which is also referred to as “nucleotide excision repair,” is a universal repair system. Excinuclease has been found in all free-living organisms, including Mycoplasma genitalium, which is considered a model system for “minimal cell.” Although both prokaryotes and eukaryotes carry out excision repair, there are distinct differences in the mechanism in the two systems.
1. Substrate
In most organisms, excision repair is the sole mechanism for repairing bulky DNA lesions. These include the vast number of base adducts formed by polyaromatic hydrocarbons and by chemotherapeutic agents such as cisplatin, mitomycin C, and psoralen, plus the cyclobutane pyrimidine dimers and the [6–4] photoproducts induced by ultraviolet light. Nucleotide excision repair is not, however, restricted to bulky adducts. Alkylated bases including O6-methylguanine and 3-methyladenine, oxidative base lesions such as 8-oxoguanine and thymine glycols, and even apurinic/apyrimidinic sites (AP sites) are removed from DNA by excision repair (5-7). Mismatched bases are also removed with low efficiency by this enzyme system; unlike mismatch repair, however, the excinuclease cannot discriminate between the correct base and the mismatched base, and this may result in fixation of a mutation (5). The most impressive aspect of the substrate repertoire of excinuclease is that it encompasses essentially all abnormal DNA structures. It must be noted, however, that, with the exception of bulky lesions, all other lesions can be repaired by alternative repair mechanisms, including repair by O6-methylguanine DNA methyltransferase, DNA glycosylases, AP endonucleases, and the mismatch repair systems. It is most likely that excision repair plays a critical backup role for these other repair systems, by repairing nonbulky DNA lesions when the normal repair enzymes are poorly expressed in a certain tissue or when a sudden burst of DNA damage exceeds the repair capacity of the specific repair mechanisms. It has been suggested that the neurological symptoms of xeroderma pigmentosum patients are caused by metabolism-induced nonbulky oxidative damage in brain cells, which overwhelms the main repair system for DNA damage caused by oxidative stress, base excision repair (7).
2. Genetics
Excision repair encompasses three basic enzymatic steps: (i) dual incisions on both sides of the damage, (ii) repair synthesis to fill in the excision gap, and (iii) ligation of the repair patch. Mutations in genes controlling any of these reactions interfere with normal functioning of excision repair. The enzymes carrying out repair synthesis and ligation are not, however, dedicated solely to repair; they also perform DNA synthesis and ligation functions during replication, recombination, and transposition. Strictly speaking, therefore, excision repair genes are the genes encoding the subunits of the excinuclease, which is the enzyme system that removes the damaged nucleotide by dual incisions bracketing the lesion. In E. coli and other prokaryotes, three genes—uvrA, uvrB, and uvrC—are necessary and sufficient for the excision reaction (3). In the yeast Saccharomyces cerevisiae, genes in the RAD3 epistasis group (RAD1, 2, 3, 4, 10, 14, 25) are required to reconstitute excinuclease (8). In humans, defects in excision repair cause xeroderma pigmentosum (XP). In this hereditary recessive disease, patients are hypersensitive to sunlight and develop skin cancers, including melanomas, at an early age. In addition, the majority of XP patients exhibit neurological signs and symptoms caused by neuronal death. There are eight XP complementation groups, XP-A through XP-G, plus the XP-variant (XP-V). Patients in complementation groups XP-A to XP-G are defective in removing pyrimidine dimers and other bulky adducts from DNA; in other words, they are defective in excision repair. In contrast, XP-V-deficient individuals exhibit normal excinuclease activity, but are defective in a process called postreplication repair. The XP complementation groups define only seven of the proteins involved in the excision reaction. In addition to these proteins, the replication protein RPA, the recombination protein ERCC1 (which makes a complex with XPF), and the transcription factor TFIIH (which includes the XPB and XPD DNA helicases and four to six additional polypeptides) are also required for dual incisions (9, 10. (
There is a one-to-one correspondence between the human and yeast excinuclease genes, and similar homologous genes have been identified in all other eukaryotes tested (11). Hence, it appears that the excision repair genes and mechanism have been conserved throughout the eukaryotic kingdom. In contrast, there is no homology between the excinuclease genes of eukaryotes and prokaryotes. Certain species in the third form of life, the Archaea, appear to have XPD, XPF, and XPG homologues, and certain others have uvrA, B, C homologues. Hence, it appears that species in this kingdom might have either the prokaryotic or eukaryotic form of excinuclease.
3. Biochemistry
The mechanism of the excision repair reaction is quite similar in prokaryotes and eukaryotes, even though there is no structural homology between the enzyme systems: Damage is recognized by the enzyme in an ATP-independent manner, forming an unstable DNA–protein complex; then ATP hydrolysis by the proteins within the complex promotes formation of a long-lived preincision complex. Binding of a nuclease subunit triggers dual incisions, which are concerted but nonsynchronous; that is, the 33′ and 5′ incisions nearly always occur in the same DNA molecule, but the 3′ incision occurs first, followed within a fraction of a second by the 5′ incision. Following both incisions, the excinuclease proteins are replaced by replication proteins. In E. coli, DNA polymerase I and DNA helicase II (UvrD protein), working together, displace the excised oligomer and the subunits of the excision nuclease that remain in the postexcision complex, and they then fill in the gap (12, 13). In yeast and humans, the replication polymerase d or , working together with the polymerase clamp loader replication factor C (RF-C) and the polymerase clamp, the proliferating cell nuclear antigen (PCNA), fill in the excision gap (14). The last step is ligation of the repair patch to the parental DNA. A more detailed description of excision repair in E. coli and humans is given below.
3.1. Excision Repair in E. coli (Fig. 1(
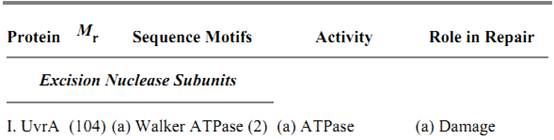
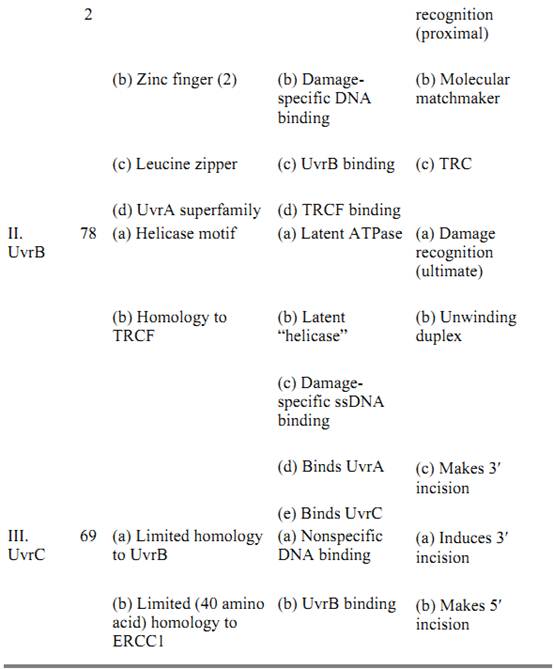
Three subunits, UvrA, UvrB, and UvrC, are necessary and sufficient to carry out the dual incision. Their properties are summarized in Table 1. UvrA and UvrB make a heterotrimer of A2B1 composition that, guided by the intrinsic affinity of UvrA for damaged DNA (15, 16), recognizes subtle or gross abnormalities in the duplex structure as damage. Upon binding to the damaged DNA, ATP hydrolysis by UvrA and UvrB leads to local unwinding of the DNA and formation of a stable UvrB–DNA complex, concomitant with dissociation of UvrA from the complex (16). Here UvrA plays the role of a molecular matchmaker (17): Utilizing the energy released by ATP hydrolysis, it brings together DNA and UvrB (a protein with rather weak affinity for DNA), promotes formation of a complex between the two, and then leaves so that the complex can engage in productive reactions. Following dissociation of UvrA, the UvrC subunit binds to the UvrB–DNA complex, and the 3′ incision is made at the fourth or fifth phosphodiester bond from the damage by UvrB, perhaps with the contribution of active-site residues of UvrC (18). After the 3′ incision, the complex undergoes another conformational change that enables UvrC, perhaps aided by residues on UvrB, to make the 5′ incision at the eighth phosphodiester bond 5′ to the lesion (18). After the dual incision, UvrB, UvrC, and the “excised” dodecamer remain in a complex with the duplex. Helicase II (UvrD) binds to this complex and releases the excised oligomer and UvrC. Then, DNA polymerase I binds to the 5′ incision site and displaces UvrB, while simultaneously filling the excision gap. When the gap is filled to the 3′ end, the repaired patch is ligated to the parental DNA. There is no nick translation in filling in the gap and, as a consequence, the repair patch size matches precisely the size of the excision gap (13).
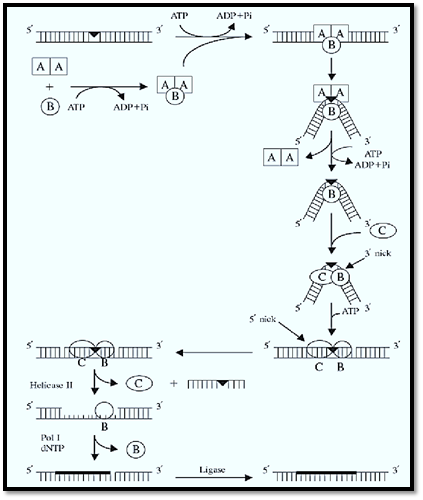
Figure 1. Excision repair in E. coli. UvrA and UvrB make up the A2B1 heterotrimer that, guided by the affinity of UvrA to helical distortion, recognizes DNA damage. Upon recognition, the DNA is locally unwound by the helicase action of the complex and kinked by 130°. Having performed its molecular matchmaking function, UvrA dissociates, leaving behind a stable UvrB–DNA complex that is recognized by UvrC. Upon binding of UvrC, UvrB makes the 3′ incision at the fifth phosphodiester bond and UvrC makes the 5′ incision at the eighth phosphodiester bond. Helicase II (UvrD) releases the excised dodecamer and UvrC; DNA pol I displaces UvrB and fills in the gap, which is ligated.
Table 1. Subunits of E. coli Excinuclease
3.2. Excision Repair in Humans (Fig. 2(
In humans, the excinuclease activity of producing dual incisions results from the coordinated action of 16 polypeptide chains in six repair factors, listed in Table 2 (9, 10). The basic reaction carried out by human excinuclease is the incision of a phosphodiester bond that is 6 ± 3 nucleotides 3′ to the damage and 20 ± 5 nucleotides 5′, followed by release of the resulting 24- to 32-nucleotide oligomers carrying the damaged base (4, 11). The reaction proceeds as follows (19): XPA, RPA, and XPC · HHR23B (20) proteins form a transient complex at the site of the damage. This complex interacts with TFIIH, which is both a transcription factor and a repair factor (21), via XPA and XPC · HHR23B and recruits this multisubunit factor to the lesion site. TFIIH unwinds the duplex by about 5 nucleotides on the 3′ side and 10 nucleotides on the 5′ side of the damaged nucleotides (19); as a consequence, it promotes new DNA–protein interactions that lead to formation of a stable preincision complex (22). XPG has high affinity for TFIIH and enters the preincision complex, either simultaneously with TFIIH or subsequently after being recruited by TFIIH, to form a very stable DNA–protein complex in which the DNA is unwound by 15 to 20 nucleotides. Entry of XPG to the preincision complex is dependent on, and concomitant with, departure of XPC from this complex. Hence, XPC plays the role of molecular matchmaker in human excinuclease. Within this complex, the 3′ incision is made by XPG (23, 24) at the sixth ± 3 phosphodiester bond 3′ to the damage. Thus, in contrast to E. coli, the 3′ incision can be made in humans before binding of the subunit that makes the 5′ incision. The XPF · ERCC1 complex is recruited to the repair assembly by the strong interaction between XPA and ERCC1, which is promoted by the presence of XPG within the complex. The XPF · ERCC1 complex makes the 5′ incision at the 20th ± 5 phosphodiester bond 5′ to the damage (24). Assembly of the excision nuclease is a highly concerted reaction, so that under physiological conditions the 3′ incision is rapidly followed by the 5′ incision; therefore, uncoupled 3′ incisions do not accumulate in the cell. The dual incision generates a single-stranded DNA fragment of 24 to 32 nucleotides, which is released from the duplex (excision) (10). This is in contrast to the bacterial system, in which the “excised” oligomer remains in the postincision complex until it is released by a DNA repair auxiliary helicase (10, 12, 13). Following release of the excised oligomer, the gap is filled by DNA polymerases d and in a reaction that is dependent on the polymerase clamp, the proliferating cell nuclear antigen (PCNA). The size of the repair patch precisely matches the size of the excision gap, with no gap enlargement on the 5′ side or nick translation on the 3′ side of the gap (25).
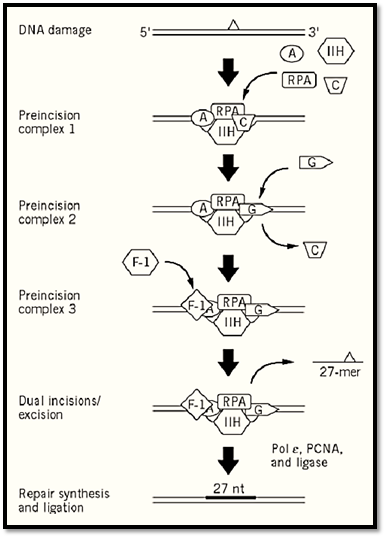
Figure 2. Excision repair in humans. XPA, RPA, XPC, and TFIIH recognize damaged DNA. TFIIH unwinds the DNA by about 20 nucleotides, and a transient DNA–protein complex forms. This structure is recognized by XPG, which enters the complex concomitant with dissociation of XPC (molecular matchmaker). XPG makes the 3′ incision at the sixth phosphodiester bond and recruits the XPF · ERCC1 complex, which makes the 5′ incision at the 22nd phosphodiester bond. The excised 27-mer is released along with some of the excinuclease subunits. The excision gap is filled in by DNA Pol Ɛ and ligated.
Table 2. Subunits of Human Excinuclease
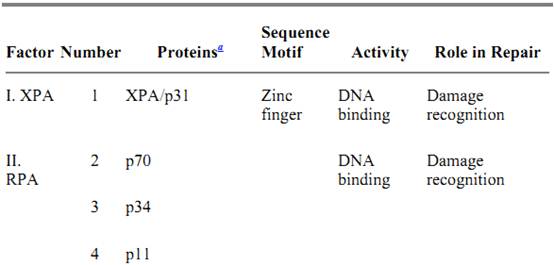
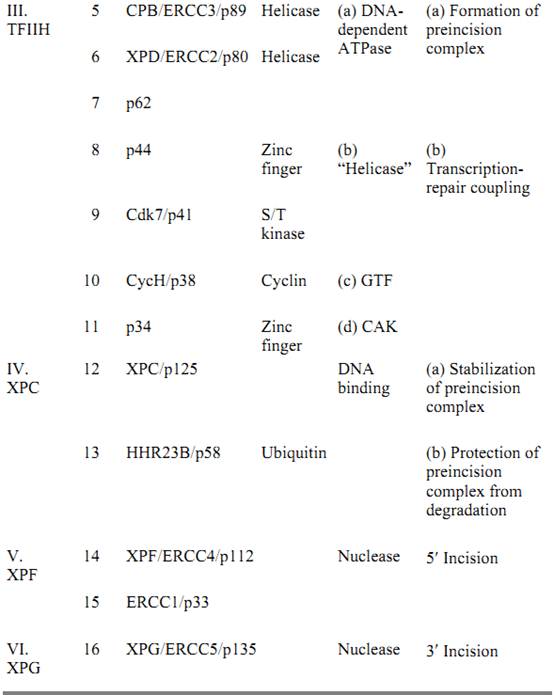
a The relative molecular weights of the polypeptides are indicated, for example, by p31 for Mr = 31 kDa, and so on. GTF, general transcription factor; CAK, cyclin-dependent kinase activating kinase.
4. Transcription-Repair Coupling
Transcribed sequences are repaired more rapidly by excinuclease than are nontranscribed sequences, both in prokaryotes and in eukaryotes (26, 27). The open chromatin conformation of transcribed DNA in eukaryotes contributes to the accessibility of the DNA to repair enzymes, and hence it has a faster rate of repair compared to regions of condensed chromatin conformation. The main driving force of preferential repair of transcribed DNA, however, is the mechanistic coupling of transcription to repair. The best manifestation of this active process of coupling is the fact that, although both strands of the DNA in the gene are more accessible because of the open conformation, the transcribed strand is repaired faster than the nontranscribed (coding) DNA strand, which is repaired at essentially the same rate as the rest of the genome (28, 29). Hence it appears that the main cause of repair coupled to transcription is the recruitment of the excinuclease complex to the site of damage (30) . Indeed, in both prokaryotes and eukaryotes, the damage-recognition step is the slowest step of excision repair. In the act of transcription, lesions in the template strand block the progression of RNA polymerase—hence the stalled complex marks the site of the lesion. In other words, RNA polymerase functions as a surrogate damage-recognition protein for lesions in the template strand. Because specific binding of repair proteins to the damage itself is very slow, due to the fact that lesions removed by excinuclease have very little in common, a stalled RNA polymerase provides a fast damage-locating mechanism and a common structural element (RNA polymerase in elongation mode) for the excision repair system. It appears both in prokaryotes and eukaryotes that a specific protein called transcription-repair coupling factor (TRCF) recognizes both the stalled RNA polymerase and a subunit of the excision nuclease and recruits the repair enzyme to the site of the lesion (30). As a result of high-specificity protein–protein interactions, rather than the low-specificity DNA–protein interaction, the rate of repair is accelerated. The transcription repair coupling factor is encoded by the mfd gene in E. coli, the RAD26 gene in yeast, and the CSB gene in humans. The molecular mechanism of transcription-repair coupling is relatively well-understood in E. coli, but not in eukaryotes.
4.1. Transcription-Repair Coupling in E. coli
The E. coli TRCF is a monomer of 130 kDa, and it has helicase motifs, a UvrA binding site in the NH 2-terminal half, and a RNAP-binding region in the middle (31). The protein recognizes RNA polymerase stalled at a lesion and displaces the stalled polymerase, while simultaneously recruiting the A2B 1 complex to the damage site. The net result is the discarding of the truncated transcript and a substantial increase in the rate of damage recognition. Furthermore, because TRCF and UvrB share the same binding site on UvrA, the TRCF also helps dissociate UvrA from the A2B1–DNA complex and facilitates rapid formation of the UvrB–DNA complex (31), which is a prerequisite for binding of UvrC. This is followed by dual incision and the remaining steps of excision repair, which are not rate-limiting. Therefore, acceleration of the damage recognition reaction and facilitation of the dissociation of UvrA by the TRCF results in overall enhancement of the excision repair rate by nearly a factor of 10 relative to excision repair unaided by RNA polymerase and TRCF. The coupling of transcription to repair results in a decline in mutation frequency of ultraviolet-irradiated cells incubated in a medium lacking amino acids prior to plating, compared to cells plated immediately on rich medium (32). In the minimal medium, the mutagenic lesions in the transcribed genes are repaired before giving rise to mutation by DNA synthesis and replication of the lesion. Hence the gene for the E. coli TRCF is called mfd (mutation frequency decline) (32).
4.2. Transcription-Repair Coupling in Humans
The phenomenology of transcription-repair coupling in humans is similar to that in E. coli, although there are some important differences. Humans defective in transcription-repair coupling are afflicted with Cockayne's syndrome, a disease characterized by growth and developmental abnormalities and mild sensitivity to sunlight. Mutations in two genes, CSA and CSB, give rise to Cockayne's syndrome. Both genes have been cloned. The CSB gene is a 160-kDa monomer with helicase motifs and appears to be the functional homologue of the mfd gene of E. coli. The CSA protein is a 55-kDa protein with the WD sequence motif, a protein–protein interaction motif. The CSB protein is a weak ATPaseand binds to RNA polymerase II either free in solution or stalled on DNA. Significantly, in contrast to the E. coli TRCF, the CSB protein, even in combination with CSA protein, does not disrupt the ternary complex of RNA polymerase II stalled at a lesion (33). CSB does bind to XPA and XPG subunits of the human excinuclease, however, and may play a role similar to that of the E. coli Mfd protein in recruiting the repair complex to the site of damage. The mechanistic details of coupling transcription to repair are not known at present.
Both in E. coli and in humans, it is the nucleotide excision repair that is tightly coupled to transcription. Recently it was found, however, that transcription also stimulates removal of thymine glycols by base excision repair in a reaction that depends on the XPG protein, but not other subunits of the human excision nuclease system. The mechanistic aspects of this coupling remain to be elucidated.
5. Regulation of Excision Repair
There is a steady-state level of damage production and removal by nucleotide excision repair under physiological conditions. It would be advantageous for the cell if, upon any sudden increase of the genotoxic load, it could increase its repair capacity. Three such regulatory systems have been characterized in E. coli: the SOS response, the adaptive response, and the adaptive response to oxidative stress. The SOS response increases the nucleotide-excision-repair capacity of the cell by increasing the level of UvrA and UvrB subunits of excinuclease. Although DNA damage induces a dramatic stress response reaction in human cells, increased repair capacity is not part of this reaction. Human excinuclease genes are not induced at the transcriptional level by DNA damage, nor is the repair activity increased as a consequence of post-transcriptional events. In particular, DNA damage causes hyperphosphorylation of the RPA p34 subunit, but this has no effect on excision repair. Similarly, certain types of damage increase the level of p53 protein by post-transcriptional mechanisms. It has been reported that this increase stimulates repair in certain cell types, but not others. The mechanism of the p53 effect on repair is not known.
References
1. R. P. Boyce and P. Howard-Flanders (1964) Proc. Natl. Acad. Sci. USA 51, 293–300.
2. R. B. Setlow and W. L. Carrier (1964) Proc. Natl. Acad. Sci. USA 51, 226–231.
3. A. Sancar and W. D. Rupp (1983) Cell 33, 249–260.
4. J. C. Huang, D. L. Svoboda, J. T. Reardon, and A. Sancar (1992) Proc. Natl. Acad. Sci. USA 89,3664-3668.
5. J. C. Huang, D. S. Hsu, A. Kazantsev, and A. Sancar (1994) Proc. Natl. Acad. Sci. USA 91, 12213-12213.
6. J. C. Huang, D. B. Zamble, J. T. Reardon, S. J. Lippard, and A. Sancar (1994) Proc. Natl. Acad. Sci. USA 91, 10394–10398.
7. J. T. Reardon, T. Bessho, H. C. Kung, P. H. Bolton, and A. Sancar (1997) Proc. Natl. Acad. Sci. USA 94, 9436–9438.
8. S. N. Guzder, Y. Habraken, P. Sung, L. Prakash, and S. Prakash (1995) J. Biol. Chem. 270, 12973-12972.
9. D. Mu, C. H. Park, T. Matsunaga, D. S. Hsu, J. T. Reardon, and A. Sancar (1995) J. Biol. Chem. 270, 2415-2418.
10. D. Mu, D. S. Hsu, and A. Sancar (1996) J. Biol. Chem. 271, 8285–8294.
11. D. L. Svoboda, J. S. Taylor, J. E. Hearst, and A. Sancar (1993) J. Biol. Chem. 268, 1931–1936.
12. P. R. Caron, S. R. Kushner, and L. Grossman (1985) Proc. Natl. Acad. Sci. USA 82, 4925–4929.
13. I. Husain, B. Van Houten, D. C. Thomas, M. Abdel-Monem, and A. Sancar (1985) Proc. Natl. Acad. Sci. USA 82, 6774–6778.
14. A. F. Nichols and A. Sancar (1992) Nucleic Acids Res. 20, 2441–2446.
15. J. T. Reardon, A. F. Nichols, S. Keeney, C. A. Smith, J. S. Taylor, S. Linn, and A. Sancar (1993) J. Biol. Chem. 268, 21301–21308.
16. D. K. Orren and A. Sancar (1989) Proc. Natl. Acad. Sci. USA 86, 5237–5241.
17. A. Sancar and J. E. Hearst (1993) Science 259, 1415–1420.
18. J. J. Lin and A. Sancar (1992) J. Biol. Chem. 267, 17688–17692.
19. M. Wakasugi and A. Sancar (1997) Proc. Natl. Acad. Sci. USA 95, 6679–6684.
20. C. Masutani, K. Sugasawa, J. Yonagisawa, T. Sanoyama, M. Ui, T. Enomoto, K. Takio, K. Tanaka, P. J. van der Spek, D. Bootsma, J. H. J. Hoeijmakers, and F. Hanaoka (1994) EMBO J. 13, 1831–1843.
21. R. Drapkin, J. T. Reardon, A. Ansari, J. C. Huang, L. Zawel, K. Ahn, A. Sancar, and D. Reinberg (1994) Nature 368, 769–772.
22. M. T. Hess, U. Schwitter, M. Petretta, B. Giese, and H. Naegeli (1997) Proc. Natl. Acad. Sci. USA 94, 6664–6669.
23. A. O''Donovan, A. A. Davis, J. A. Moggs, S. C. West, and R. D. Wood (1994) Nature 371, 432–435.
24. T. Matsunaga, C. H. Park, T. Bessho, D. Mu, and A. Sancar (1996) J. Biol. Chem. 271, 11047–11050.
25. J. T. Reardon, L. H. Thompson, and A. Sancar (1997) Nucleic Acids Res. 25, 1015–1021.
26. R. C. Bockrath and J. E. Palmer (1997) Mol. Gen. Genet. 156, 133–140.
27. V. A. Bohr, C. A. Smith, D. S. Okumoto, and P. C. Hanawalt (1985) Cell 40, 359–369.
28. I. Mellon and P. C. Hanawalt (1989) Nature 342, 95–98.
29.I. Mellon, G. Spirak, and P. C. Hanawalt (1987) Cell 51, 241–249.
30. C. P. Selby and A. Sancar (1993) Science 260, 53–58.
31. C. P. Selby and A. Sancar (1995) J. Biol. Chem. 270, 4882–4889.
32. E. M. Witkin (1966) Science 152, 1345–1353.
33. C. P. Selby and A. Sancar (1997) J. Biol. Chem. 272, 1885–1890.



|
|
|
|
للعاملين في الليل.. حيلة صحية تجنبكم خطر هذا النوع من العمل
|
|
|
|
|
|
|
"ناسا" تحتفي برائد الفضاء السوفياتي يوري غاغارين
|
|
|
|
|
|
|
نحو شراكة وطنية متكاملة.. الأمين العام للعتبة الحسينية يبحث مع وكيل وزارة الخارجية آفاق التعاون المؤسسي
|
|
|