


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 2-11-2020
Date: 2-5-2016
Date: 31-5-2021
|
DNA Methylation Is Responsible for Imprinting
KEY CONCEPTS
- Paternal and maternal alleles may have different patterns of methylation at fertilization.
- Methylation is usually associated with inactivation of the gene.
- When genes are differentially imprinted, survival of the embryo may depend on whether a functional allele is provided by the parent with the unmethylated allele.
- Survival of heterozygotes for imprinted genes is different, depending on the direction of the cross.
- Imprinted genes occur in clusters and may depend on a local control site where de novo methylation occurs unless specifically prevented.
The pattern of methylation of germ cells is established in each sex during gametogenesis by a two-stage process: First, the existing pattern is erased by a genome-wide demethylation in primordial germ cells and then a pattern specific for each sex is imposed during meiosis.
All allelic differences are lost when primordial germ cells develop in the embryo; irrespective of sex, the previous patterns of methylation are erased, and a typical gene is then unmethylated. In males, the pattern develops in two stages. The methylation pattern that is characteristic of mature sperm is established in the spermatocyte, but further changes are made in this pattern after fertilization. In females, the maternal pattern is imposed during oogenesis, when oocytes mature through meiosis after birth.
As may be expected from the inactivity of genes in gametes, the typical state is to be methylated. Some cases of differences between the two sexes have been identified, though, for which a locus is unmethylated in one sex. A major question is how the specificity of methylation is determined in the male and female gametes.
Systematic changes occur in early embryogenesis. Some sites will continue to be methylated, whereas others will be specifically unmethylated in cells in which a gene is expressed. From the pattern of changes, it may be inferred that individual sequencespecific demethylation events occur during somatic development of the organism as particular genes are activated.
The specific pattern of DNA methylation in germ cells is responsible for the phenomenon of imprinting, which describes a difference in behavior between the alleles inherited from each parent. The expression of certain genes in mouse embryos (and other mammals) depends upon the sex of the parent from which they were inherited. For example, the allele encoding insulin-like growth factor II (IGF-II) that is inherited from the father is expressed, but the allele that is inherited from the mother is not expressed. The IGF-II gene of oocytes is methylated in its promoter, whereas the IGF-II gene of sperm is not, so that the two alleles behave differently in the zygote. This is the most common pattern, but the dependence on sex is reversed for some genes. In fact, the opposite pattern (expression of maternal copy) is shown for IGFIIR, a gene encoding a receptor that causes the rapid turnover of IGF-II.
This sex-specific mode of inheritance requires that the pattern of methylation be established specifically during each gametogenesis. The fate of a hypothetical locus in a mouse is illustrated in FIGURE 1. In the early embryo, the paternal allele is unmethylated and expressed, and the maternal allele is methylated and silent. What happens when this mouse itself forms gametes? If it is a male, the allele contributed to the sperm must be nonmethylated, irrespective of whether it was originally methylated or not. Thus, when the maternal allele finds itself in a sperm, it must be demethylated. If the mouse is a female, the allele contributed to the egg must be methylated; if it was originally the paternal allele, methyl groups must be added.
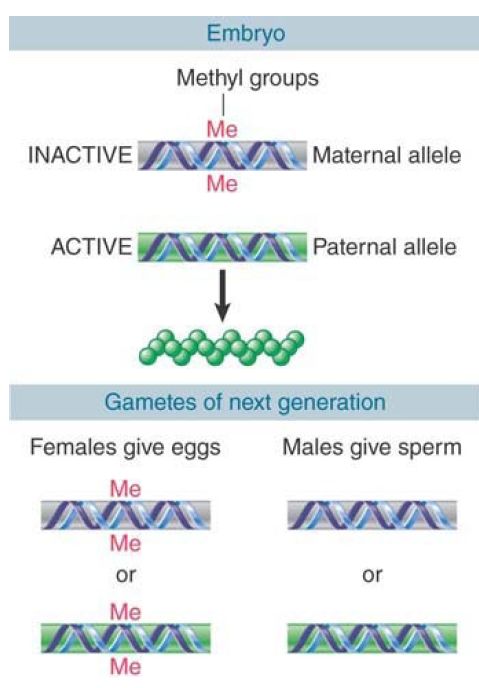
FIGURE 1. The typical pattern for imprinting is that a methylated locus is inactive. If this is the maternal allele, only the paternal allele is active, and it may be essential for viability. The methylation pattern is reset when gametes are formed so that all sperm have the paternal type and all oocytes have the maternal type.
The consequence of imprinting is that an embryo is hemizygous for any imprinted gene. Thus, in the case of a heterozygous cross where the allele of one parent has an inactivating mutation, the embryo will survive if the wild-type allele comes from the parent in which this allele is active but will die if the wild-type allele is the imprinted (silenced) allele. This type of dependence on the directionality of the cross (in contrast with Mendelian genetics) is an example of epigenetic inheritance, where some factor other than the sequences of the genes themselves influences their effects.
Although the paternal and maternal alleles can have identical sequences, they display different properties, depending on which parent provided them. These properties are inherited through meiosis and the subsequent somatic mitoses. Although imprinted genes are estimated to comprise 1% to 2% of the mammalian transcriptome, these genes are sometimes clustered. More than half of the 25 or so known imprinted genes in mice are contained in six particular regions, each containing both maternally and paternally expressed genes. This suggests the possibility that imprinting mechanisms may function over long distances. Some insights into this possibility come from deletions in the human population that cause Prader–Willi and Angelman syndromes. Most cases of these neurodevelopmental disorders involving the proximal long arm of chromosome 15 are caused by the same 4-Mb deletion, but the syndromes are different, depending on which parent contributed the deletion. The reason is that the deleted region contains at least one gene that is paternally imprinted and at least one that is maternally imprinted. Thus, affected individuals receive one chromosome missing a given allele due to the deletion, and the corresponding (intact) allele from the other parent is imprinted and thus silent. This results in affected individuals being functionally null for these alleles.
In some rare cases, however, affected individuals present with much smaller deletions. Prader–Willi syndrome can be caused by a 20-kb deletion that silences distant genes on either side of the deletion. The basic effect of the deletion is to prevent a father from resetting the paternal mode to a chromosome inherited from his mother. The result is that these genes remain in maternal mode so that both the paternal and maternal alleles are silent in the offspring. The inverse effect is found in some small deletions that cause Angelman syndrome. These mutations have led to the identification of a Prader–Willi/Angelman syndrome “imprint center” (PW/AS IC) that acts at a distance to regulate imprinting in either sex across the entire region.
A microdeletion resulting in removal of a cluster of small nucleolar RNAs (snoRNAs) that is paternally derived may result in the key aspects of Prader–Willi syndrome. Mutations that separate the snoRNA HBII-85 cluster from its promoter cause Prader–Willi syndrome, although other genes in the region could also contribute to the syndrome.
Six imprinted regions are often associated with disease in humans, and the phenotypic diversity of these disorders is related to the multiple genes in the imprinted regions. These defects in imprinted genes may take the form of aberrant expression involving loss or overexpression of genes. For example, in Russell–Silver syndrome, an overexpression of maternal alleles and loss of paternal gene expression for chromosome 11p15.5 result in this syndrome that is characterized by an undergrowth disorder.
Imprinting may also regulate alternative polyadenylation. A number of mammalian genes utilize multiple polyadenylation (polyA) sites to confer diversity on gene transcription. The H13 murine gene undergoes alternative polyadenylation in an allele-specific manner, in that polyA sites are differentially methylated in the maternal and paternal genome of this imprinted gene. Elongation proceeds to downstream polyadenylation sites when the allele is methylated,indicating that epigenetic processes may influence alternative polyadenylation, contributing to the diversity of gene transcription in mammals.



|
|
|
|
التوتر والسرطان.. علماء يحذرون من "صلة خطيرة"
|
|
|
|
|
|
|
مرآة السيارة: مدى دقة عكسها للصورة الصحيحة
|
|
|
|
|
|
|
نحو شراكة وطنية متكاملة.. الأمين العام للعتبة الحسينية يبحث مع وكيل وزارة الخارجية آفاق التعاون المؤسسي
|
|
|