


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 24-12-2015
Date: 2025-02-27
Date: 30-11-2015
|
The Lambda Repressor Uses a Helix-Turn-Helix Motif to Bind DNA
KEY CONCEPTS
- Each DNA-binding region in the repressor contacts a half-site in the DNA.
- The DNA-binding site of the repressor includes two short α-helical regions that fit into the successive turns of the major groove of DNA.
- A DNA-binding site is a (partially) palindromic sequence of 17 bp.
- The amino acid sequence of the recognition helix makes contact with particular bases in the operator sequence that it recognizes.
A repressor dimer is the unit that binds to DNA. It recognizes a sequence of 17 bp displaying partial symmetry about an axis through the central base pair. FIGURE 1 shows an example of a binding site. The sequence on each side of the central base pair is sometimes called a half-site. Each individual N-terminal region contacts a half-site. Several DNA-binding proteins that regulate bacterial transcription share a similar mode of holding DNA, in which the active domain contains two short regions of α-helix that contact DNA. (Some transcription factors in eukaryotic cells use a similar motif).

FIGURE 1.The operator is a 17-bp sequence with an axis of symmetry through the central base pair. Each half-site is marked in light blue. Base pairs that are identical in each operator half are in dark blue.
The N-terminal domain of lambda repressor contains several stretches of α-helix, which are arranged as illustrated diagrammatically in FIGURE 2. Two of the helical regions are responsible for binding DNA. The helix-turn-helix model for contact is illustrated in FIGURE 3. Looking at a single monomer, α-helix-3 consists of nine amino acids, each of which lies at an angle to the preceding region of seven amino acids that forms α-helix-2. In the dimer, the two apposed helix-3 regions lie 34 Å apart, enabling them to fit into successive major grooves of DNA. The helix-2 regions lie at an angle that would place them across the groove. The symmetrical binding of dimer to the site means that each N-terminal domain of the dimer contacts a similar set of bases in its half-site.
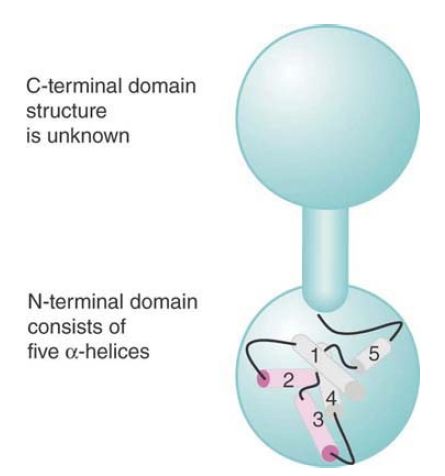
FIGURE 2.Lambda repressor’s N-terminal domain contains five stretches of α-helix; helices 2 and 3 bind DNA.
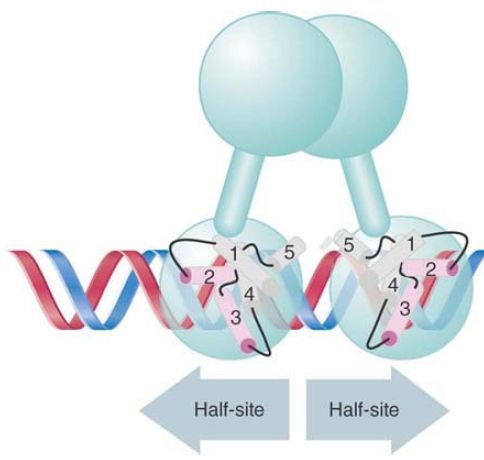
FIGURE 3. In the two-helix model for DNA binding, helix-3 of each monomer lies in the wide groove on the same face of DNA and helix-2 lies across the groove.
Related forms of the α-helical motifs employed in the helix-turnhelix of the lambda repressor are found in several DNA-binding proteins, including catabolite repressor protein (CRP), the lac repressor, and several other phage repressors. By comparing the abilities of these proteins to bind DNA, the roles of each helix can be defined:
- Contacts between helix-2 and helix-3 are maintained by interactions between hydrophobic amino acids.
- Contacts between helix-3 and DNA rely on hydrogen bonds between the amino acid side chains and the exposed positions of the base pairs. This helix is responsible for recognizing the
specific target DNA sequence and is therefore also known as the recognition helix. Comparison of the contact patterns illustrated in FIGURE 4shows that the lambda repressor and Cro select different sequences in the DNA as their most favored targets because they have different amino acids in the corresponding positions in helix-3.
- Contacts from helix-2 to the DNA take the form of hydrogen bonds connecting with the phosphate backbone. These interactions are necessary for binding, but do not control the specificity of target recognition. In addition to these contacts, a large part of the overall energy of interaction with DNA is provided by ionic interactions with the phosphate backbone.

FIGURE 4. Two proteins that use the two-helix arrangement to contact DNA recognize lambda operators with affinities determined by the amino acid sequence of helix-3.
What happens if we manipulate the coding sequence to construct a new protein by substituting the recognition helix in one repressor with the corresponding sequence from a closely related repressor? The specificity of the hybrid protein is that of its new recognition helix. The amino acid sequence of this short region determines the sequence specificities of the individual proteins and is able to act in conjunction with the rest of the polypeptide chain.
The bases contacted by helix-3 lie on one face of the DNA, as can be seen from the positions indicated on the helical diagram in Figure 4. Repressor makes an additional contact with the other face of DNA, though. The last six N-terminal amino acids of the N-terminal domain form an “arm” extending around the back. FIGURE 5 shows the view from the back. Lysine residues in the arm make contact with G residues in the major groove, and also with the phosphate backbone. The interaction between the arm and DNA contributes heavily to DNA binding; the binding affinity of a mutant armless repressor is reduced by about 1,000-fold.

FIGURE 5. A view from the back shows that the bulk of the repressor contacts one face of DNA, but its N-terminal arms reach around to the other face.



|
|
|
|
التوتر والسرطان.. علماء يحذرون من "صلة خطيرة"
|
|
|
|
|
|
|
مرآة السيارة: مدى دقة عكسها للصورة الصحيحة
|
|
|
|
|
|
|
نحو شراكة وطنية متكاملة.. الأمين العام للعتبة الحسينية يبحث مع وكيل وزارة الخارجية آفاق التعاون المؤسسي
|
|
|