


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 10-12-2015
Date: 8-3-2021
Date: 7-11-2020
|
Modified Bases Affect Anticodon–Codon Pairing
KEY CONCEPT
- Modifications in the anticodon affect the pattern of wobble pairing and therefore are important in determining tRNA specificity.
tRNA modifications in and adjacent to the anticodon influence its ability to pair with the mRNA codon. Most such modifications are present at positions 34 and 37 of the anticodon loop, and they generally function by constraining the range of available motion in the anticodon. In turn, this facilitates docking of the tRNA into the A site of the ribosome. These modifications influence codon pairing, and as a result they directly function to help determine how the cell assigns the meaning of the tRNA. Modified bases permit further pairing patterns in addition to those involving regular and wobble pairing of A, C, U, and G.
Inosine is particularly important when present at the first anticodon position (nucleotide 34 in the sequence) because it is able to pair with any one of the three bases U, C, or A (FIGURE 1). The role of inosine is well illustrated in the decoding of isoleucine codons. Here AUA encodes isoleucine, whereas AUG encodes methionine. To read the A at the third codon position, a tRNA would require U at the first anticodon position—but this U in the wobble position would necessarily also pair with G. Thus any tRNA with a 5′ U in its anticodon would recognize both AUG and AUA. This problem is resolved by synthesis of an isoleucine tRNA possessing A34, followed by modification of A34 to I34 by the enzyme tRNA adenosine deaminase. I34 then is able to recognize all three codons of the isoleucine set: AUU, AUC, and AUA.
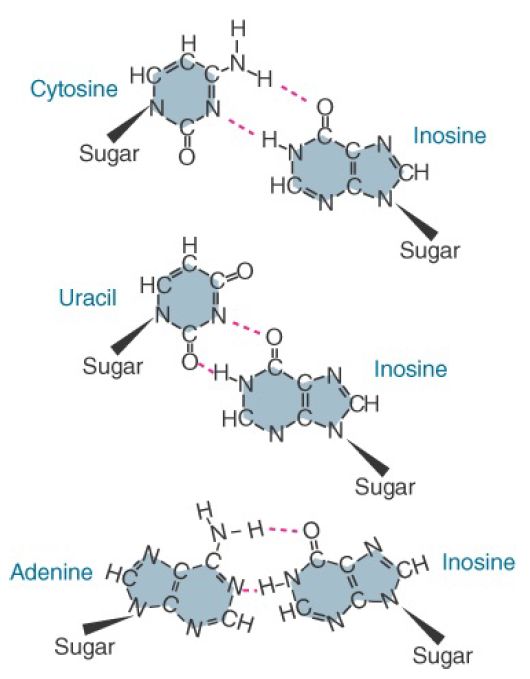
FIGURE 1. Inosine can pair with U, C, or A.
In most cases, U at the first position of the anticodon is also converted to a modified form that has altered pairing properties. Derivatives of U possessing the 2-thio group in place of oxygen show improved selectivity in pairing to A as compared with G (FIGURE 2). Anticodons with uridine-5-oxyacetic acid and related modifications in the first position have the remarkable property of permitting the single tRNA to read three and sometimes all four of the synonymous codons NNA, NNC, NNU, and NNG.

FIGURE 2. Modification to 2-thiouridine restricts pairing to A alone because only one H-bond can form with G.
These and other pairing relationships show that there are multiple ways to construct a set of tRNAs able to recognize all the 61 codons representing amino acids. No particular pattern predominates in any particular organism, although the absence of a certain pathway for modification can prevent the use of some recognition patterns. Thus, a particular codon family is read by tRNAs with different anticodons in different organisms.
Often the tRNAs will have overlapping capacities to read certain codons, so that a particular codon is read by more than one tRNA. In such cases there may be differences in the efficiencies of the alternative recognition reactions. (As a general rule, codons that are commonly used tend to be more efficiently read.)
The predictions of wobble pairing accord very well with experimental evidence for almost all tRNAs. However, exceptions exist in which the codons recognized by a tRNA differ from those predicted by the wobble rules. Such effects probably result from the influence of neighboring bases and/or the conformation of the anticodon loop in the overall tertiary structure of the tRNA. Further support for the influence of the surrounding structure is provided by the isolation of occasional mutants in which a change in a base in some other region of the molecule alters the ability of the anticodon to recognize codons.



|
|
|
|
5 علامات تحذيرية قد تدل على "مشكل خطير" في الكبد
|
|
|
|
|
|
|
لحماية التراث الوطني.. العتبة العباسية تعلن عن ترميم أكثر من 200 وثيقة خلال عام 2024
|
|
|