

علم الكيمياء

تاريخ الكيمياء والعلماء المشاهير

التحاضير والتجارب الكيميائية

المخاطر والوقاية في الكيمياء

اخرى

مقالات متنوعة في علم الكيمياء

كيمياء عامة


الكيمياء التحليلية

مواضيع عامة في الكيمياء التحليلية

التحليل النوعي والكمي

التحليل الآلي (الطيفي)

طرق الفصل والتنقية


الكيمياء الحياتية

مواضيع عامة في الكيمياء الحياتية

الكاربوهيدرات

الاحماض الامينية والبروتينات

الانزيمات

الدهون

الاحماض النووية

الفيتامينات والمرافقات الانزيمية

الهرمونات


الكيمياء العضوية

مواضيع عامة في الكيمياء العضوية

الهايدروكاربونات

المركبات الوسطية وميكانيكيات التفاعلات العضوية

التشخيص العضوي

تجارب وتفاعلات في الكيمياء العضوية


الكيمياء الفيزيائية

مواضيع عامة في الكيمياء الفيزيائية

الكيمياء الحرارية

حركية التفاعلات الكيميائية

الكيمياء الكهربائية


الكيمياء اللاعضوية

مواضيع عامة في الكيمياء اللاعضوية

الجدول الدوري وخواص العناصر

نظريات التآصر الكيميائي

كيمياء العناصر الانتقالية ومركباتها المعقدة


مواضيع اخرى في الكيمياء

كيمياء النانو

الكيمياء السريرية

الكيمياء الطبية والدوائية

كيمياء الاغذية والنواتج الطبيعية

الكيمياء الجنائية


الكيمياء الصناعية

البترو كيمياويات

الكيمياء الخضراء

كيمياء البيئة

كيمياء البوليمرات

مواضيع عامة في الكيمياء الصناعية

الكيمياء الاشعاعية والنووية
Translation
المؤلف:
William Reusch
المصدر:
Virtual Textbook of Organic Chemistry
الجزء والصفحة:
............
27-8-2018
3268
Translation
Translation is a more complex process than transcription. This would, of course, be expected. After all, the coded messages produced by the German Enigma machine could be copied easily, but required a considerable decoding effort before they could be read with understanding. In a similar sense, DNA replication is simply a complementary base pairing exercise, but the translation of the four letter (bases) alphabet code of RNA to the twenty letter (amino acids) alphabet of protein literature is far from trivial. Clearly, there could not be a direct one-to-one correlation of bases to amino acids, so the nucleotide letters must form short words or codons that define specific amino acids. Many questions pertaining to this genetic code were posed in the late 1950's:
• How many RNA nucleotide bases designate a specific amino acid?
If separate groups of nucleotides, called codons, serve this purpose, at least three are needed. There are 43 = 64 different nucleotide triplets, compared with 42 = 16 possible pairs.
• Are the codons linked separately or do they overlap?
Sequentially joined triplet codons will result in a nucleotide chain three times longer than the protein it describes. If overlapping codons are used then fewer total nucleotides would be required.
• If triplet segments of mRNA designate specific amino acids in the protein, how are the codons identified?
For the sequence ~CUAGGU~ are the codons CUA & GGU or ~C, UAG & GU~ or ~CU, AGG & U~?
• Are all the codon words the same size?
In Morse code the most widely used letters are shorter than less common letters. Perhaps nature employs a similar scheme.
Physicists and mathematicians, as well as chemists and microbiologists all contributed to unravelling the genetic code. Although earlier proposals assumed efficient relationships that correlated the nucleotide codons uniquely with the twenty fundamental amino acids, it is now apparent that there is considerable redundancy in the code as it now operates. Furthermore, the code consists exclusively of non-overlapping triplet codons.
Clever experiments provided some of the earliest breaks in deciphering the genetic code. Marshall Nirenberg found that RNA from many different organisms could initiate specific protein synthesis when combined with broken E.coli cells (the enzymes remain active). A synthetic polyuridine RNA induced synthesis of poly-phenylalanine, so the UUU codon designated phenylalanine. Likewise an alternating ~CACA~ RNA led to synthesis of a ~His-Thr-His-Thr~ polypeptide.
The following table presents the present day interpretation of the genetic code. Note that this is the RNA alphabet, and an equivalent DNA codon table would have all the U nucleotides replaced by T. Methionine and tryptophan are uniquely represented by a single codon. At the other extreme, leucine is represented by eight codons. The average redundancy for the twenty amino acids is about three. Also, there are three stop codons that terminate polypeptide synthesis.
|
Second Position |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U | C | A | G | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
F |
U |
|
|
|
|
|
T |
||||||||||||||||||||||||||||||||||||||||||||||||||||
| C |
|
|
|
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| A |
|
|
|
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| G |
|
|
|
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
The translation process is fundamentally straightforward. The mRNA strand bearing the transcribed code for synthesis of a protein interacts with relatively small RNA molecules (about 70-nucleotides) to which individual amino acids have been attached by an ester bond at the 3'-end.
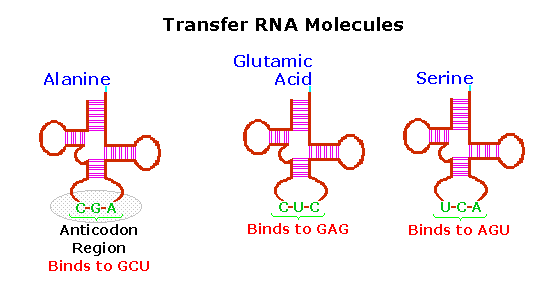
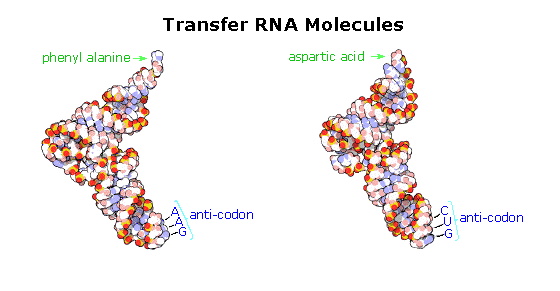
These transfer RNA's (tRNA) have distinctive three-dimensional structures consisting of loops of single-stranded RNA connected by double stranded segments. This cloverleaf secondary structure is further wrapped into an "L-shaped" assembly, having the amino acid at the end of one arm, and a characteristic anti-codon region at the other end. The anti-codon consists of a nucleotide triplet that is the complement of the amino acid's codon(s). Models of two such tRNA molecules are shown to the right. When read from the top to the bottom, the anti-codons depicted here should complement a codon in the previous table.
Cloverleaf cartoons of three other tRNA molecules will be shown on the right by clicking on the diagram.
A cell's protein synthesis takes place in organelles called ribosomes. Ribosomes are complex structures made up of two distinct and separable subunits (one about twice the size of the other). Each subunit is composed of one or two RNA molecules (60-70%) associated with 20 to 40 small proteins (30-40%). The ribosome accepts a mRNA molecule, binding initially to a characteristic nucleotide sequence at the 5'-end (colored light blue in the following diagram). This unique binding assures that polypeptide synthesis starts at the right codon. A tRNA molecule with the appropriate anti-codon then attaches at the starting point and this is followed by a series of adjacent tRNA attachments, peptide bond formation and shifts of the ribosome along the mRNA chain to expose new codons to the ribosomal chemistry.
The following diagram is designed as a slide show illustrating these steps. The outcome is synthesis of a polypeptide chain corresponding to the mRNA blueprint. A "stop codon" at a designated position on the mRNA terminates the synthesis by introduction of a "Release Factor".
 الاكثر قراءة في الاحماض النووية
الاكثر قراءة في الاحماض النووية
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















