


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 28-11-2015
Date: 25-2-2021
Date: 9-12-2015
|
Databases
Molecular biology is an empirical discipline that requires observing and understanding different types of data. For example, a gene may imply a physical location on the chromosome, a nucleotide sequence, an amino acid sequence, a three-dimensional protein structure, a molecular component of cellular function, a regulatory mechanism of gene expression, or even a phenotypic difference caused by mutation. In addition to this variability, the quantity of molecular biology data is increasing rapidly, especially for gene and protein sequences and 3-D structures, due to advances in experimental technologies. Molecular biology databases are a number of resources available over the Internet that comprise a bioinformatics infrastructure for biomedical sciences. Each database contains a specific type of data that has cross-references to other databases, which can be used for integrated information retrieval. This is possible because a database is generally organized as a collection of entries, and connections can be made at the level of entries without standardizing how data items should be organized within an entry. The advent of the World Wide Web (WWW) in the early 1990s was a boon to molecular biology databases because the concept of hyperlinks is fully compatible with the practice of cross-references. The WWW also dramatically increased the accessibility of computers to biologists. The primary resources of molecular biology databases are bibliographic databases, sequence databases, and structure databases.
Although bibliographic, sequence, and structural aspects of molecular biology are relatively easy to computerize, the next step is to organize their functional aspects. There are resources in that direction, such as motif libraries that contain higher level knowledge abstracted from sets of functionally related sequences and pathway databases that contain computerized knowledge of molecular interactions and biochemical pathways. Representative examples of these resources are shown in Table 1.
Table 1. Selected List of Molecular Biology Databases
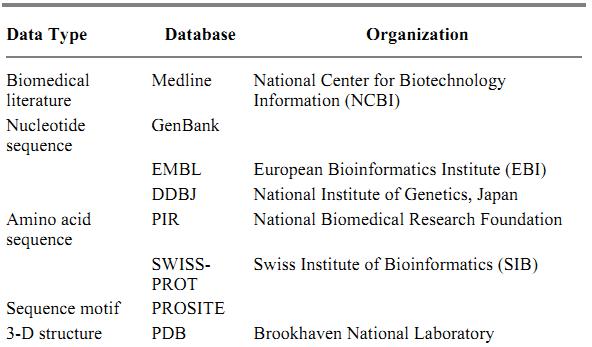
Abstraction of a real problem is made through a data model. For example, data are organized in two-dimensional tables in the relational data model. The relational database based on the relational model has been widely used in a number of applications, including some of the sequence databases. Although in principle all different types of molecular biology data can be stored in a single, unified, relational database, this is impossible in practice because of the varying views of how data items should be organized and related. In the current web of molecular biology databases, different types of data are integrated by a loose coupling based on links (cross-references), rather than a tight coupling based on unified schema. This approach is extended to include other types of links, especially similarity links computed by similarity search algorithms and biological links representing molecular interactions, which can also be integrated for biological reasoning (1). The major bioinformatics servers shown in Table 2 provide link-based database retrieval systems, such as Entrez at NCBI, SRS at EBI, and DBGET/LinkDB at GenomeNet in Kyoto.
In scientific disciplines, the merit of storing and managing information in a computer was first realized in bibliographic databases, which were designed for humans to read and understand. In the next step, factual data reported in the literature were computerized in factual databases, such as in sequence databases, which made more sophisticated retrieval available, for example, sequence similarity searches. Even in this case, however, the database is still a static resource to be retrieved, and it is up to humans to make sense out of the retrieved data. In contrast, the knowledge of links or relationships is more dynamic in nature. For example, ancestors in a family can be retrieved from a “deductive” database that contains parent–child relationships and the rules for combining them. Thus, knowledge is different from data or information, in that new knowledge can be generated dynamically from existing knowledge by logical reasoning. In the era of mass data production, molecular biology requires logical computation based on empirical knowledge rather than numerical computation based on first principles. The web of molecular biology databases also requires a new generation of knowledge bases.
References
1. M. Kanehisa (1997) Trends Biochem. Sci. 22, 442–444.



|
|
|
|
لخفض ضغط الدم.. دراسة تحدد "تمارين مهمة"
|
|
|
|
|
|
|
طال انتظارها.. ميزة جديدة من "واتساب" تعزز الخصوصية
|
|
|
|
|
|
|
أعمال المؤتمر العلمي السنوي الدولي الرابع عشر بعنوان .. "الإمامان الحسنان عليهما السلام صراطُ حقٍ ومشكاةُ معرفةٍ"
|
|
|