
Regulation of Eukaryotic Gene Expression: Regulation through variations in DNA
 المؤلف:
Denise R. Ferrier
المؤلف:
Denise R. Ferrier
 المصدر:
Lippincott Illustrated Reviews: Biochemistry
المصدر:
Lippincott Illustrated Reviews: Biochemistry
 الجزء والصفحة:
الجزء والصفحة:
 31-12-2021
31-12-2021
 1263
1263
Regulation of Eukaryotic Gene Expression: Regulation through variations in DNA
Gene expression in eukaryotes is also influenced by the accessibility of DNA to the transcriptional apparatus, the amount of DNA, and the arrangement of DNA. [Note: Localized transitions between the B and Z forms of DNA can also affect gene expression.]
1. Access to DNA:
In eukaryotes, DNA is found complexed with histone and nonhistone proteins to form chromatin . Transcriptionally active, decondensed chromatin (euchromatin) differs from the more condensed, inactive form (heterochromatin) in a number of ways. Active chromatin contains histone proteins that have been covalently modified at their amino terminal ends by reversible methylation, acetylation, or phosphorylation . Such modifications decrease the positive charge of these basic proteins, thereby decreasing the strength of their association with negatively charged DNA. This relaxes the nucleosome , allowing transcription factors access to specific regions on the DNA.
Nucleosomes can also be repositioned, an ATP-requiring process that is part of chromatin remodeling. Another difference between transcriptionally active and inactive chromatin is the extent of methylation of cytosine bases in CG-rich regions (CpG islands) in the promoter region of many genes. Methylation is by methyltransferases that use S-adenosylmethionine as the methyl donor (Fig. 1).
Transcriptionally active genes are less methylated (hypomethylated) than their inactive counterparts, suggesting that DNA hypermethylation silences gene expression. Modification of histones and methylation of DNA are epigenetic in that they are heritable changes in DNA that alter gene expression without altering the base sequence.
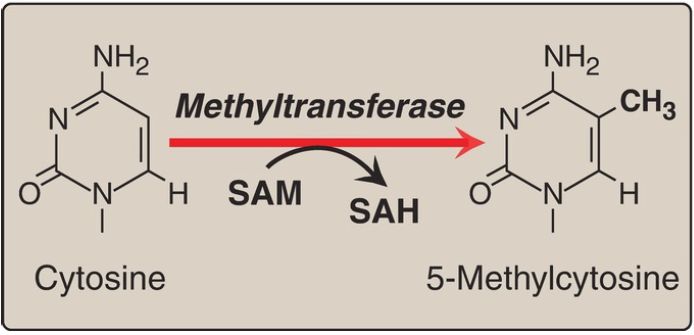
Figure 1: The methylation of cytosine in eukaryotic DNA. SAM = Sadenosylmethionine; SAH = S-adenosylhomocysteine.
2. Amount of DNA:
A change up or down in the number of copies of a gene can affect the amount of gene product produced. An increase in copy number (gene amplification) has contributed to increased genomic complexity and is still a normal developmental process in certain nonmammalian species. In mammals, however, gene amplification is seen with some diseases and in response to particular chemotherapeutic drugs such as methotrexate, an inhibitor of the enzyme dihydrofolate reductase (DHFR), required for the synthesis of thymidine triphosphate (TTP) in the pyrimidine biosynthetic pathway . TTP is essential for DNA synthesis. Gene amplification results in an increase in the number of DHFR genes and resistance to the drug, allowing TTP to be made.
3. Arrangement of DNA:
The process by which immunoglobulins (antibodies) are produced by B lymphocytes involves permanent rearrangements of the DNA in these cells. The immunoglobulins (for example, IgG) consist of two light and two heavy chains, with each chain containing regions of variable and constant amino acid sequence. The variable region is the result of somatic recombination of segments within both the light- and the heavy-chain genes. During B-lymphocyte development, single variable (V), diversity (D), and joining (J) gene segments are brought together through gene rearrangement to form a unique variable region (Fig. 2). This process allows the generation of 109−1011 different immunoglobulins from a single gene, providing the diversity needed for the recognition of an enormous number of antigens. [Note: Pathologic DNA rearrangement is seen with translocation, a process by which two different chromosomes exchange DNA segments.]
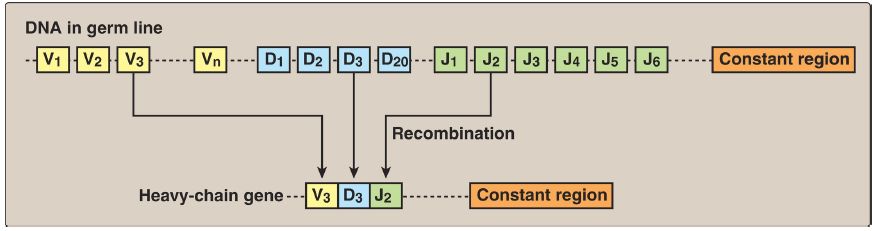
Figure 2: DNA rearrangements in the generation of immunoglobulins. V= variable; D = diversity; J = joining.
4. Mobile DNA elements:
Transposons (Tn) are mobile segments of DNA that move in an essentially random manner from one site to another on the same or a different chromosome. Movement is mediated by transposase, an enzyme encoded by the Tn itself. Movement can be direct, in which transposase cuts out and then inserts the Tn at a new site, or replicative, in which the Tn is copied and the copy inserted elsewhere while the original remains in place. In eukaryotes, including humans, replicative transposition frequently involves an RNA intermediate made by a reverse transcriptase , in which case the Tn is called a retrotransposon. Transposition has contributed to structural variation in the genome but also has the potential to alter gene expression and even to cause disease. Tn comprise ~50% of the human genome, with retrotransposons accounting for 90% of Tn. Although the vast majority of these retrotransposons have lost the ability to move, some are still active. Their transposition is thought to be the basis for some rare cases of hemophilia A and Duchenne muscular dystrophy.
[Note: The growing problem of antibiotic-resistant bacteria is a consequence, at least in part, of the exchange of plasmids among bacterial cells. If the plasmids contain Tn-carrying antibiotic resistance genes, the recipient bacteria gain resistance to one or more antimicrobial drugs.]
 الاكثر قراءة في الكيمياء الحيوية
الاكثر قراءة في الكيمياء الحيوية
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


