

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Map for Protein Synthesis
المؤلف:
Denise R. Ferrier
المصدر:
Lippincott Illustrated Reviews: Biochemistry
الجزء والصفحة:
29-12-2021
1579
Map for Protein Synthesis
Codons are composed of three nucleotide bases presented in the messenger RNA (mRNA) language of adenine (A), guanine (G), cytosine (C), and uracil (U). They are always written 5′→3′. Of the 64 possible three-base combinations, 61 code for the 20 standard amino acids and 3 signal termination of protein synthesis (translation). Altering the nucleotide sequence in a codon can cause silent mutations (the altered codon codes for the original amino acid), missense mutations (the altered codon codes for a different amino acid), or nonsense mutations (the altered codon is a termination codon). Characteristics of the genetic code include specificity, universality, and degeneracy, and it is nonoverlapping and commaless (Fig.1). Requirements for protein synthesis include all the amino acids that eventually appear in the finished protein; at least one specific type of transfer RNA (tRNA) for each amino acid; one aminoacyl-tRNA synthetase for each amino acid; the mRNA coding for the protein to be synthesized; fully competent ribosomes (70S in prokaryotes, 80S in eukaryotes); protein factors needed for initiation, elongation, and termination of protein synthesis; and ATP and guanosine triphosphate (GTP) as energy sources. tRNA has an attachment site for a specific amino acid at its 3′-end and an anticodon region that can recognize the codon specifying the amino acid the tRNA is carrying. Ribosomes are large complexes of protein and ribosomal RNA (rRNA).
They consist of two subunits, 30S and 50S in prokaryotes and 40S and 60S in eukaryotes. Each ribosome has three binding sites for tRNA molecules: the A, P, and E sites that cover three neighboring codons. The A site binds an incoming aminoacyl-tRNA, the P site is occupied by peptidyl-tRNA, and the E site is occupied by the empty tRNA as it is about to exit the ribosome.
Recognition of an mRNA codon is accomplished by the tRNA anticodon, which binds to the codon following the rules of complementarity and antiparallel binding. The wobble hypothesis states that the first (5′) base of the anticodon is not as spatially defined as the other two bases. Movement of that first base allows nontraditional base-pairing with the last (3′) base of the codon, thus allowing a single tRNA to recognize more than one codon for a specific amino acid. For initiation of protein synthesis, the components of the translation system are assembled, and mRNA associates with the small ribosomal subunit. The process requires initiation factors (IF). In prokaryotes, a purine-rich region of the mRNA (the Shine-Dalgarno sequence) base-pairs with a complementary sequence on 16S rRNA, resulting in the positioning of the small subunit on the mRNA so that translation can begin. The 5′-cap (bound by proteins of the eIF-4 family) on eukaryotic mRNA is used to position the small subunit on the mRNA. The initiation codon is AUG, and N-formylmethionine is the initiating amino acid in prokaryotes, whereas methionine is used in eukaryotes. The charged initiating tRNA (tRNAi) is brought to the P site by (e)IF-2. In elongation, the polypeptide chain is lengthened by the addition of amino acids to the carboxyl end of its growing chain. The process requires elongation factors that facilitate the binding of the aminoacyl-tRNA to the A site as well as the movement of the ribosome along the mRNA. The formation of the peptide bond is catalyzed by peptidyltransferase, which is an activity intrinsic to the rRNA of the large subunit and, therefore, is a ribozyme.
Following peptide-bond formation, the ribosome advances along the mRNA in the 5′→3′ direction to the next codon (translocation). Because of the length of most mRNA, more than one ribosome at a time can translate a message, forming a polysome. Termination begins when one of the three termination codons moves into the A site. These codons are recognized by release factors. The newly synthesized protein is released from the ribosomal complex, and the ribosome is dissociated from the mRNA.
Initiation, elongation, and termination are driven by the hydrolysis of GTP. Initiation in eukaryotes also requires ATP for scanning. Numerous antibiotics interfere with the process of protein synthesis. Many polypeptide chains are covalently modified during or after translation. Such modifications include amino acid removal; phosphorylation, which may activate or inactivate the protein; glycosylation, which plays a role in protein targeting; and hydroxylation such as that seen in collagen. Protein targeting can be either cotranslational (as with secreted proteins) or posttranslational (as with mitochondrial matrix proteins). Proteins must fold to achieve their functional form. Folding can be spontaneous or facilitated by chaperones. Proteins that are defective (for example, misfolded) or destined for rapid turnover are marked for destruction by the attachment of chains of a small, highly conserved protein called ubiquitin. Ubiquitinated proteins are rapidly degraded by a cytosolic complex known as the proteasome.
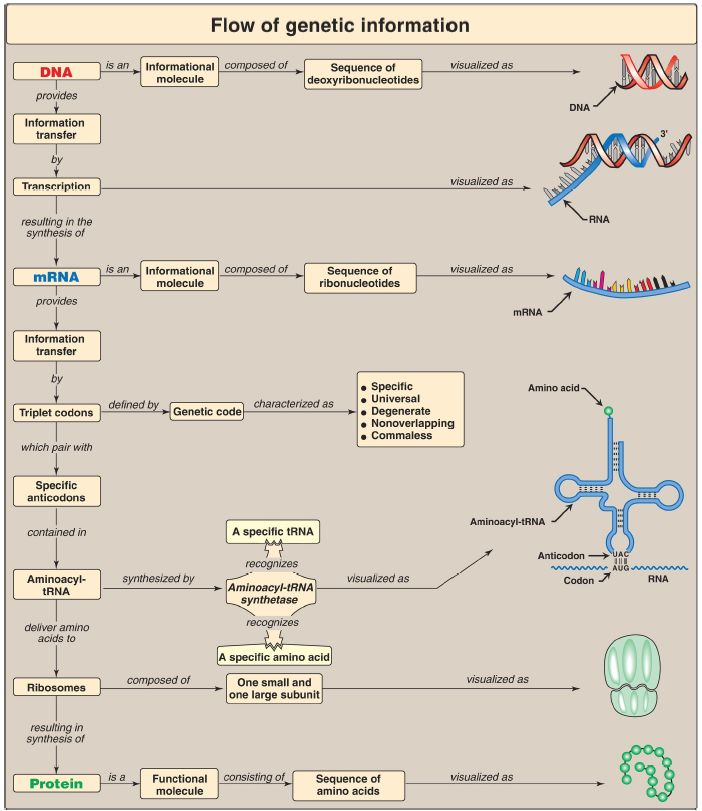
Figure 1: Key concept map for protein synthesis. mRNA = messenger RNA; tRNA = transfer RNA; A = adenine; G = guanine; C = cytosine; U = uracil.
 الاكثر قراءة في الكيمياء الحيوية
الاكثر قراءة في الكيمياء الحيوية
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















