
Introduction to The Operon
 المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
 المصدر:
LEWIN’S GENES XII
المصدر:
LEWIN’S GENES XII
 الجزء والصفحة:
الجزء والصفحة:
 2-6-2021
2-6-2021
 3599
3599
Introduction to The Operon
KEY CONCEPTS
- In negative regulation, a repressor protein binds to an operator to prevent a gene from being expressed.
- In positive regulation, a transcription factor is required to bind at the promoter to enable RNA polymerase to initiate transcription.
- In inducible regulation, the gene is regulated by the presence of its substrate.
- In repressible regulation, the gene is regulated by the product of its enzyme pathway.
- Gene regulation in vivo can utilize any of these mechanisms, resulting in four combinations: negative inducible, negative repressible, positive inducible, and positive repressible.
Gene expression can be controlled at any of several stages, which can be divided broadly into transcription, processing, and translation:
- Transcription often is controlled at the stage of initiation.
- Transcription is not usually controlled at elongation, but it may be controlled at termination to determine whether RNA polymerase is allowed to proceed past a terminator to the gene(s) beyond.
- In bacteria, an mRNA is typically available for translation while it is being synthesized; this is called coupled transcription/translation. (In eukaryotic cells, transcription takes place in the nucleus, and translation takes place in the cytoplasm.)
- Translation in bacteria may be directly regulated, but more commonly it is passively modulated. The coding portion or open reading frame of a gene can be assembled either with common or rare codons, which correspond to common or rare tRNAs. mRNAs containing a number of rare codons take longer to translate.
The basic concept for the way transcription is controlled in bacteria is called the operon model and was proposed by François Jacob and Jacques Monod in 1961. They distinguished between two types of sequences in DNA: sequences that code for trans-acting products (usually proteins) and cis-acting DNA sequences. Gene activity is regulated by the specific interactions of the trans-acting products with the cis-acting sequences . In more formal terms:
- A gene is a sequence of DNA that codes for a diffusible product, either RNA or a protein. The crucial feature is that the product diffuses away from its site of synthesis to act elsewhere. Any gene product that is free to diffuse to find its target is described as trans-acting.
- The description cis-acting applies to any sequence of DNA that functions exclusively as a DNA sequence, affecting only the DNA to which it is physically linked.
To help distinguish between the components of regulatory circuits and the genes that they regulate, the terms structural gene and regulator gene are sometimes used. A structural gene is simply any gene that codes for a protein (or RNA) product. Protein structural genes represent an enormous variety of structures and functions, including structural proteins, enzymes with catalytic activities, and regulatory proteins. One type of structural gene is a regulator gene, which is simply a gene that codes for a protein or an RNA involved in regulating the expression of other genes.
The simplest form of the regulatory model is illustrated in FIGURE 1: A regulator gene codes for a protein that controls transcription by binding to particular site(s) on DNA. This interaction can regulate a target gene in either a positive manner (the interaction turns the gene on) or a negative manner (the interaction turns the gene off). The sites on DNA are usually (but
not exclusively) located just upstream of the target gene.
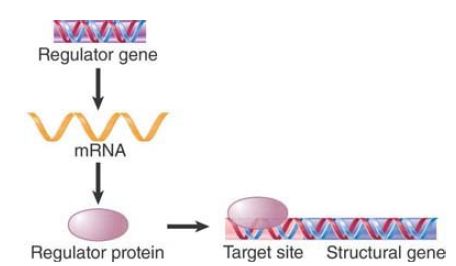
FIGURE .1 A regulator gene codes for a protein that acts at a target site on DNA.
The sequences that mark the beginning and end of the transcription unit—the promoter and terminator—are examples of cis-acting sites. A promoter serves to initiate transcription only of the gene(s) physically connected to it on the same stretch of DNA. In the same way, a terminator can terminate transcription only by an RNA polymerase that has traversed the preceding gene(s). In their simplest forms, promoters and terminators are cis-acting elements that are recognized by the same trans-acting species; that is, by RNA polymerase (although other factors also participate at each site).
Additional cis-acting regulatory sites are often combined with the promoter. A bacterial promoter may have one or more such sites located close by; that is, in the immediate vicinity of the start point. A eukaryotic promoter is likely to have a greater number of sites that are spread out over a longer distance, as described in the chapter titled Eukaryotic Transcription Regulation.
A classic mode of transcription control in bacteria is negative control: A repressor protein prevents a gene from being expressed. FIGURE .2 shows that in the absence of the negative regulator the gene is expressed. Close to the promoter is another cis-acting site called the operator, which is the binding site for the repressor protein. When the repressor binds to the operator, RNA polymerase is prevented from initiating transcription, and gene expression is therefore turned off. An alternative mode of control is positive control. This is used in bacteria (probably) with about equal frequency to negative control, and it is the most common mode of control in eukaryotes. A transcription factor is required to assist RNA polymerase in initiating at the promoter.
FIGURE .3 shows that in the absence of the positive regulator the gene is inactive: RNA polymerase cannot by itself initiate transcription at the promoter.
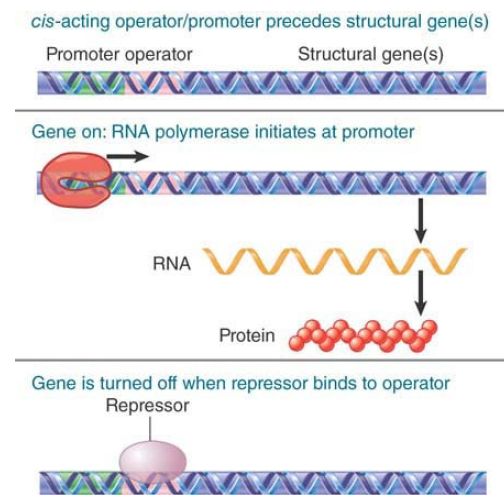
FIGURE.2 In negative control, a trans-acting repressor binds to the cis-acting operator to turn off transcription.

FIGURE .3 In positive control, a trans-acting factor must bind to the cis-acting site in order for RNA polymerase to initiate transcription at the promoter.
In addition to negative and positive control, a gene that encodes an enzyme may be regulated by the concentration of its substrate or product (or a chemical derivative of either). Bacteria need to respond swiftly to changes in their environment. Fluctuations in the supply of nutrients (such as the sugars glucose or lactose) can occur at any time, and survival depends on the ability to switch from metabolizing one substrate to another. Yet economy is important, too: A bacterium that indulges in energetically expensive ways to meet the demands of the environment is likely to be at a disadvantage. Thus, a bacterium avoids synthesizing the enzymes of a pathway in the absence of the substrate, but is ready to produce the enzymes if the substrate should appear. The synthesis of enzymes in response to the appearance of a specific substrate is called induction and the gene is an inducible gene.
The opposite of induction is repression, where the repressible gene is controlled by the amount of the product made by the enzyme. For example, Escherichia coli synthesizes the amino acid tryptophan through the actions of an enzyme complex containing tryptophan synthetase and four other enzymes. If, however, tryptophan is provided in the medium on which the bacteria are growing, the production of the enzyme is immediately halted. This allows the bacterium to avoid devoting its resources to unnecessary synthetic activities.
Induction and repression represent similar phenomena. In one case the bacterium adjusts its ability to use a given substrate (such as lactose) for growth; in the other it adjusts its ability to synthesize a particular metabolic intermediate (such as an essential amino acid). The trigger for either type of adjustment is a small molecule that is the substrate (or related to the substrate) for the enzyme or the product of the enzyme activity, respectively. Small molecules that cause the production of enzymes that are able to metabolize them (or their analogues) are called inducers. Those that prevent the production of enzymes that are able to synthesize them are called corepressors.
These two ways of looking at regulation—negative versus positive control and inducible versus repressible control—are typically combined to give four different patterns of gene regulation: negative inducible, negative repressible, positive inducible, and positive repressible, as shown in FIGURE 4. This enables a bacterium to perform the ultimate in inventory control of its metabolism to allow survival in rapidly changing environments.
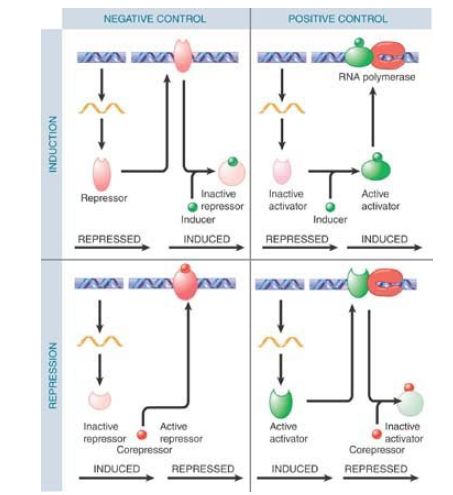
FIGURE 4. Regulatory circuits can be designed from all possible combinations of positive and negative control with inducible and repressible control.
The unifying theme is that regulatory proteins are trans-acting factors that recognize cis-acting elements (usually) upstream of the gene. The consequences of this recognition are either to activate or to repress the gene, depending on the individual type of regulatory protein. A typical feature is that the protein functions by recognizing a very short sequence in DNA, usually less than 10 bp in length, although the protein actually binds over a somewhat greater distance of DNA. The bacterial promoter is an example: RNA polymerase covers less than 70 bp of DNA at initiation, but the crucial sequences that it recognizes are the hexamers centered at −35 and −10.
A significant difference in gene organization between prokaryotes and eukaryotes is that structural genes in bacteria are organized in operons that are coordinately controlled by means of interactions at a single regulator. In contrast, genes in eukaryotes are usually controlled individually. As a result, an entire related set of bacterial genes is either transcribed or not transcribed.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


