
mRNA-Specific Half-Lives Are Controlled by Sequences or Structures Within the mRNA
 المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
 المصدر:
LEWIN’S GENES XII
المصدر:
LEWIN’S GENES XII
 الجزء والصفحة:
الجزء والصفحة:
 19-5-2021
19-5-2021
 2273
2273
mRNA-Specific Half-Lives Are Controlled by Sequences or Structures Within the mRNA
KEY CONCEPTS
- Specific cis-elements in an mRNA affect its rate of degradation.
- Destabilizing elements (DEs) can accelerate mRNA decay, whereas stabilizing elements (SEs) can reduce it.
- AU-rich elements (AREs) are common destabilizing elements in mammals and are bound by a variety of proteins.
- Some DE-binding proteins interact with components of the decay machinery and probably recruit them for degradation.
- Stabilizing elements occur on some highly stable mRNAs.
- mRNA degradation rates can be altered in response to a variety of signals.
What accounts for the large range of half-lives of different mRNAs in the same cell? Specific cis-elements within an mRNA are known to affect its stability. The most common location for such elements is within the 3′ UTR, although they exist elsewhere. Whole-genome studies have revealed many highly conserved 3′ UTR motifs, but their roles remain mostly unknown. Many are likely to be target sites for miRNA base pairing. Others are binding sites for RBPs,some of which have known functions in stability. Rates of deadenylation can vary widely for different mRNAs, and sequences that affect this rate have been described.
Destabilizing elements (DEs) have been the most widely studied. The criterion for defining a destabilizing sequence element is that its introduction into a more stable mRNA accelerates its degradation. Removal of an element from an mRNA does not necessarily stabilize it, indicating that an individual mRNA can have more than one DE. To complicate their identification further, the presence of a DE does not guarantee a short half-life under all conditions, because other sequence elements in the mRNA can modify its effectiveness.
The most well-studied type of DE is the AU-rich element (ARE), found in the 3′ UTR of up to 8% of mammalian mRNAs. AREs are heterogeneous, and a number of subtypes have been characterized. One type consists of the pentamer sequence AUUUA present once or repeated multiple times in different sequence contexts. Another type does not contain AUUUA and is predominantly U-rich. A large number of ARE-binding proteins with specificity for certain ARE types and/or cell types have been identified. How do AREs work to stimulate rapid degradation?
Many ARE-binding proteins have been found to interact with one or more components of the degradation machinery, including the exosome, deadenylases, and decapping enzyme, suggesting that they act by recruiting the degradation machinery. The exosome can bind some AREs directly. The AREs of a number of mRNAs have been shown to accelerate the deadenylation step of decay, although it is not likely that they all work this way. Another way they might act is by facilitating efficient engagement of the mRNA into processing bodies.
Many AU-rich DEs and other kinds of destabilizing elements have been identified in the mRNAs of budding yeast and other model organisms. For example, the previously mentioned Puf proteins of yeast bind to specific UG-rich elements and accelerate the degradation of target mRNAs. In this case, the destabilizing mechanism is accelerated deadenylation by recruitment of the CCR4-NOT deadenylase. A genomics analysis of yeast 3′ UTRs has identified 53 sequence elements that correlate with the halflives of mRNAs containing them, suggesting the number of different destabilizing elements may be large. FIGURE 1 summarizes the known actions of destabilizing elements.
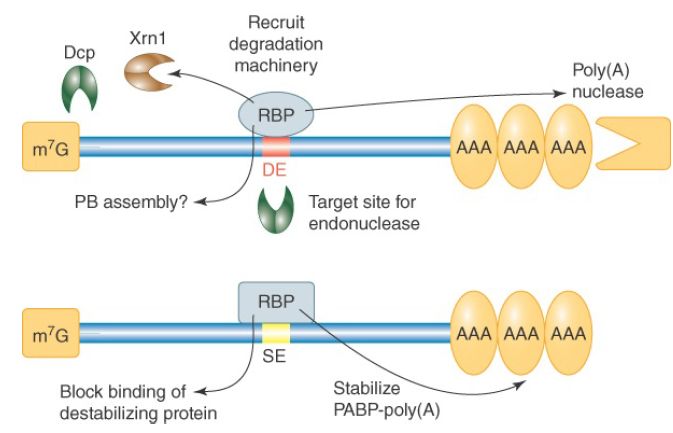
FIGURE 1. Mechanisms by which destabilizing elements (DEs) and stabilizing elements (SEs) function. Effects of DEs and SEs on mRNA stability are mediated primarily through the proteins that bind to them. One exception is a DE that acts as an endonuclease target site.
Stabilizing elements (SEs) have been identified in a few unusually stable mRNAs. Three mRNAs studied in mammalian cells have stabilizing pyrimidine-rich sequences in their 3′ UTRs. Proteins that bind to this element in globin mRNA have been shown to interact with PABPs, suggesting they might function to protect the poly(A) tail from degradation. In some cases, an mRNA can be stabilized by inhibition of its DE. For example, certain ARE-binding proteins act to prevent the ARE from destabilizing the mRNA, presumably by blocking the ARE-binding site. An example of regulated mRNA stabilization occurs for the mammalian transferrin mRNA. It is stabilized when its 3′ UTR iron-response element (IRE), consisting of multiple stem-loop structures, is bound by a specific protein, as shown in FIGURE 2. The affinity of the IRE-binding protein for the IRE is altered by iron binding, exhibiting low affinity when its iron-binding site is full and high affinity when it is not. When the cellular iron concentration is low, more transferrin is needed to import iron from the bloodstream, and under these conditions the transferrin mRNA is stabilized. The IRE-binding protein stabilizes the mRNA by inhibiting the function of destabilizing sequences in the vicinity. Interestingly, the same IRE-binding protein also binds an IRE in ferritin mRNA and regulates this mRNA in a very different way. Ferritin is an iron-binding protein that sequesters excess cellular iron. The IRE-binding protein binds IRE stem-loops in the 5′ UTR of ferritin when iron is low and blocks the interaction of the cap-binding complex with ferritin mRNA. Thus, translation of ferritin mRNA is prevented when cellular iron levels are low—the conditions under which transferrin mRNA is stabilized and translated.
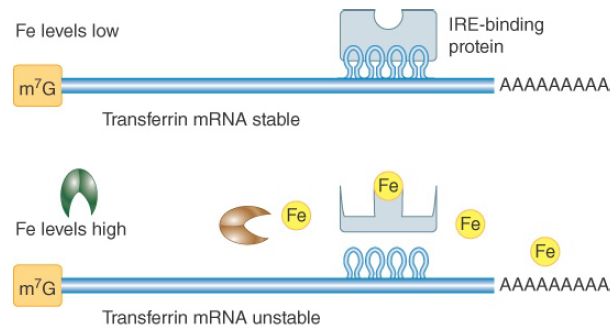
FIGURE 2.Regulation of transferring mRNA stability by iron (Fe) levels. The IRE in the 3′ UTR is the binding site for a protein that stabilizes the mRNA. The IRE-binding protein is sensitive to iron levels in the cell, binding to the IRE only when iron is low.
Many cis-element–binding proteins are subject to modifications that are likely to affect their functions, including phosphorylations, methylations, conformational changes due to effector binding, and isomerizations. Such modifications may be responsible for changes in mRNA degradation rates induced by cellular signals. mRNA decay can be altered in response to a wide variety of environmental and internal stimuli, including cell cycle progression, cell differentiation, hormones, nutrient supply, and viral infection.
Microarray studies have shown that almost 50% of changes in mRNA levels stimulated by cellular signals are due to mRNA stabilization or destabilization events, not to transcriptional changes. How these changes are effected remains largely unknown.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


