

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
LINEs Use an Endonuclease to Generate a Priming End
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
28-4-2021
2620
LINEs Use an Endonuclease to Generate a Priming End
KEY CONCEPT
- LINEs do not have LTRs and require the retroposon to code for an endonuclease that generates a nick to prime reverse transcription.
LINEs, like all retroposons, do not terminate in the LTRs that are typical of retroviral elements. This poses the question: How is reverse transcription primed? It does not involve the typical reaction, in which a tRNA primer pairs with the LTR. The open reading frames in these elements lack many of the retroviral functions, such as protease or integrase domains, but typically have reverse transcriptase–like sequences and code for an endonuclease activity. In the human LINE L1, ORF1 is a DNAbinding protein and ORF2 has both reverse transcriptase and endonuclease activities; both products are required for transposition.
FIGURE 1 shows how these activities support transposition. A nick is made in the DNA target site by an endonuclease activity encoded by the retroposon. The RNA product of the element associates with the protein bound at the nick. The nick provides a 3′–OH end that primes synthesis of cDNA on the RNA template. A second cleavage event is required to open the other strand of DNA, and the RNA–DNA hybrid is linked to the other end of the gap either at this stage or after it has been converted into a DNA duplex. A similar mechanism is used by some mobile introns .
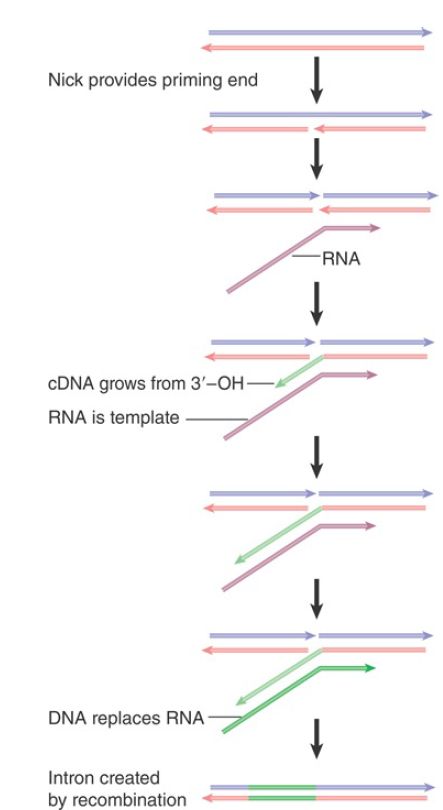
FIGURE 1. Retrotransposition of non-LTR retroposons occurs by nicking the target to provide a primer for cDNA synthesis on an RNA template. The arrowheads indicate 3′ ends.
One of the reasons why LINEs are so effective lies with their method of propagation. When a LINE mRNA is translated, the protein products show a cis-preference for binding to the mRNA from which they were translated. FIGURE 2 shows that the ribonucleoprotein complex then moves to the nucleus, where the proteins insert a DNA copy into the genome. Reverse transcription often does not proceed fully to the end, resulting in a truncated and inactive element. The potential exists, however, for insertion of an active copy, because the proteins are acting in cis on a transcript of the original active element.
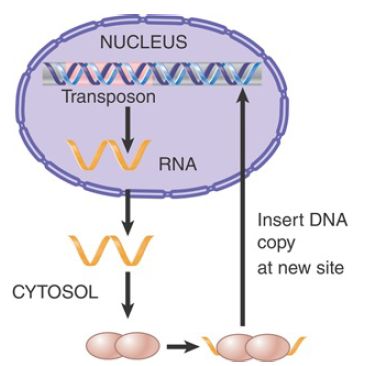
FIGURE 2. A LINE is transcribed into an RNA that is translated into proteins that assemble into a complex with the RNA. The complex translocates to the nucleus, where it inserts a DNA copy into the genome.
By contrast, the proteins produced by the DNA transposons must be imported into the nucleus after being synthesized in the cytoplasm, but they have no means of distinguishing full-length transposons from inactive deleted transposons. FIGURE 3 shows that instead of distinguishing these two types of transposons, the proteins will indiscriminately recognize any element by virtue of the repeats that mark the ends. This greatly reduces their chance of acting on a full-length element as opposed to one that has been deleted, resulting in an inability to replicate the autonomous elements efficiently. This can potentially lead to extinction of the entire family of elements.
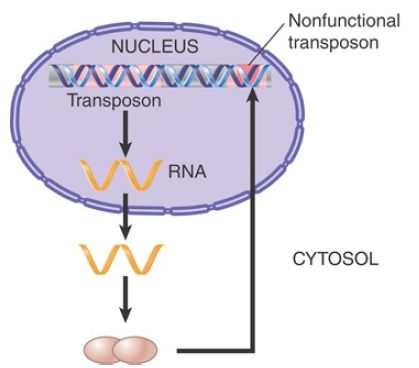
FIGURE 3. A transposon is transcribed into an RNA that is translated into proteins that move independently to the nucleus, where they act on any pair of inverted repeats with the same sequence as the original transposon.
Are transposition events of retroelements currently occurring in these genomes, or are we seeing only the footprints of ancient systems? This varies with the species. Only a few transposons are currently active in the human genome, but several active transposons are known in the mouse genome. This explains the fact that spontaneous mutations caused by LINE insertions occur at a rate of about 3% in mice, but only 0.1% in humans. It appears that 80 to 100 LINEs are active in the human genome. Some human diseases can be pinpointed as the result of transposition of L1 into genes, and others result from unequal crossing-over events
involving repeated copies of L1. A model system in which LINE transposition occurs in tissue culture cells suggests that a transposition event can introduce several types of collateral damage as well as inserting into a new site; the damage includes chromosomal rearrangements and deletions. Such events may be viewed as agents of genetic change. Neither DNA transposons nor retroviral-like retrotransposons seem to have been active in the human genome for 40 to 50 million years, but several active examples of both are found in the mouse.
Note that for transpositions to survive, they must occur in the germline. Similar events occur in somatic cells, but do not survive beyond one generation.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















