

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
The Bacterial RecBCD System Is Stimulated by chi Sequences
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
15-4-2021
3088
The Bacterial RecBCD System Is Stimulated by chi Sequences
KEY CONCEPTS
- The RecBCD complex has nuclease and helicase activities.
- RecBCD binds to DNA downstream of a chi sequence, unwinds the duplex, and degrades one strand from 3′→5′ as it moves to the chi site.
- The chi site triggers loss of the RecD subunit and nuclease activity.
The nature of the events involved in exchange of sequences between DNA molecules was first described in bacterial systems. Here the recognition reaction is part and parcel of the recombination mechanism and involves restricted regions of DNA molecules rather than intact chromosomes. The general order of molecular events is similar, though: A single strand from a broken molecule interacts with a partner duplex, the region of pairing is extended, and an endonuclease resolves the partner duplexes.
Enzymes involved in each stage are known, although they probably represent only some of the components required for recombination. Bacterial enzymes implicated in recombination have been identified by the occurrence of rec mutations in their genes. The phenotype of rec mutants is the inability to undertake generalized recombination. Some 10 to 20 loci have been identified. Bacteria do not usually exchange large amounts of duplex DNA, but there may be various routes to initiate recombination in prokaryotes. In some cases, DNA may be available with free single-stranded 3′ ends: DNA may be provided in single-stranded form , single-stranded gaps may be generated by irradiation damage, or single-stranded tails may be generated by phage genomes undergoing replication by a rolling circle. In circumstances involving two duplex molecules (as in recombination at meiosis in eukaryotes), however, single-stranded regions and 3′ ends must be generated.
One mechanism for generating suitable ends has been discovered as a result of the existence of certain hotspots that stimulate recombination. These hotspots, which were discovered in phage lambda in the form of mutants called chi, have single base–pair changes that create sequences that stimulate recombination. These sites lead us to the role of other proteins involved in recombination.
These sites share a constant nonsymmetrical sequence of 8 bp:
5′ GCTGGTGG 3′
3′ CGACCACC 5′
The chi sequence occurs naturally in E. coli DNA about once every 5 to 10 kb. Its absence from wild-type lambda DNA, and also from other genetic elements, shows that it is not essential for recombination.
A chi sequence stimulates recombination in its general vicinity, within about a distance of up to 10 kb from the site. A chi site can be activated by a DSB made several kilobases away on one particular side (to the right of the sequence shown previously). This dependence on orientation suggests that the recombination apparatus must associate with DNA at a broken end, and then can move along the duplex only in one direction.
chi sites are targets for the action of an enzyme encoded by the genes recBCD. This complex possesses several activities: It is a potent nuclease that degrades DNA (originally identified as the activity exonuclease V); it has helicase activities that can unwind duplex DNA in the presence of a single-strand binding (SSB) protein; and it has ATPase activity. Its role in recombination may be to provide a single-stranded region with a free 3′ end.
FIGURE 1 shows how these reactions are coordinated on a substrate DNA that has a chi site. RecBCD binds to DNA at a double-stranded end. Two of its subunits have helicase activities: RecD functions with 5′→3′ polarity, and RecB functions with 3′→5′ polarity. Translocation along DNA and unwinding the double helix is initially driven by the RecD subunit. As RecBCD advances, it degrades the released single strand with the 3′ end. When it reaches the chi site, it recognizes the top strand of the chi site in single-stranded form. This causes the enzyme to pause. It then cleaves the top strand of the DNA at a position between four and six bases to the right of chi. Recognition of the chi site causes the RecD subunit to dissociate or become inactivated, at which point the enzyme loses its nuclease activity. It continues, however, to function as a helicase—now using only the RecB subunit to drive translocation—at about half the previous speed. The overall result of this interaction is to generate single-stranded DNA with a 3′ end at the chi sequence. This is a substrate for recombination.
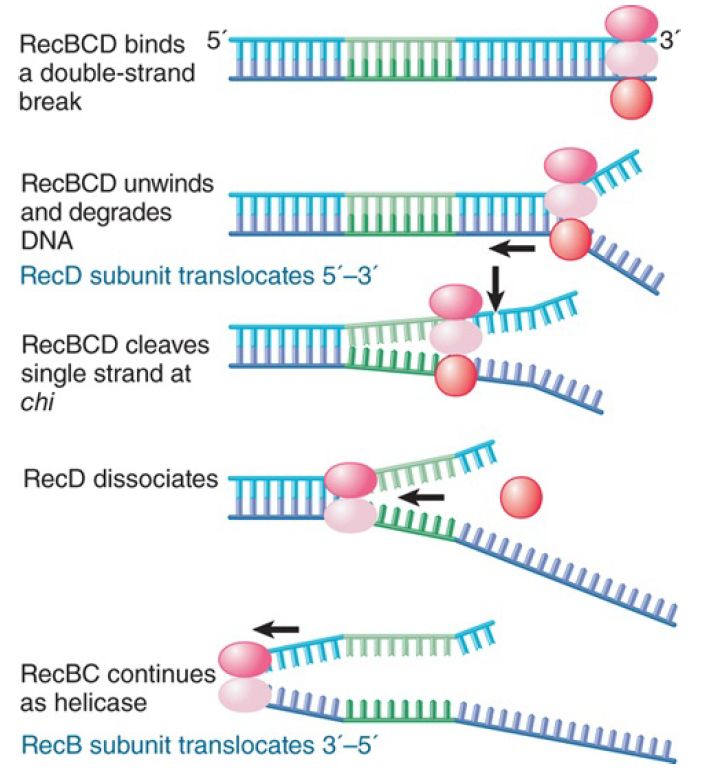
FIGURE 1. RecBCD nuclease approaches a chi sequence from one side, degrading DNA as it proceeds; at the chi site, it makes an endonucleolytic cut, loses RecD, and retains only the helicase activity.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















