

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الحيوية

مواضيع عامة في المضادات الحيوية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
The Synaptonemal Complex Forms After Double-Strand Breaks
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
15-4-2021
1854
The Synaptonemal Complex Forms After Double-Strand Breaks
KEY CONCEPTS
- Double-strand breaks that initiate recombination occur before the synaptonemal complex forms.
- If recombination is blocked, the synaptonemal complex cannot form.
- Meiotic recombination involves two phases: one that results in gene conversion without crossover, and one that results in crossover products.
Evidence suggests that DSBs initiate recombination in both homologous and site-specific recombination in yeast. DSBs were initially implicated in the change of mating type, which involves the replacement of one sequence by another . DSBs also occur early in meiosis at sites
that provide hotspots for recombination. Their locations are not sequence specific. They tend to occur in promoter regions and to coincide with more accessible regions of chromatin. The frequency of recombination declines in a gradient on one or both sides of the hotspot. The hotspot identifies the site at which recombination is initiated, and the gradient reflects the probability that the recombination events will spread from it.
We may now interpret the role of DSBs in molecular terms. The blunt ends created by the DSB are rapidly converted on both sides into long 3′–single-stranded ends, A yeast mutation (rad50) that blocks the conversion of the blunt end into the single-stranded protrusion is defective in recombination. This suggests that DSBs are necessary for recombination. The gradient is determined by the declining probability that a single-stranded region will be generated as distance increases from the site of the DSB.
In rad50 mutants, the 5′ ends of the DSBs are connected to the protein Spo11, which, as discussed previously, is homologous to the catalytic subunits of a family of type II topoisomerases. Spo11 generates the DSBs. Recall that the model for this reaction, , suggests that Spo11 interacts reversibly with DNA; the break is converted into a permanent structure by an interaction with another protein that dissociates the Spo11 complex. Removal of Spo11 is then followed by nuclease action. At least nine other proteins are required to process the DSBs. One group of proteins is required to convert the DSBs into protruding 3′–OH singlestranded ends. Another group then enables the single- stranded ends to invade homologous duplex DNA.
The correlation between recombination and synaptonemal complex formation is well established in most species, and recent work has shown that all mutations that abolish chromosome pairing in Drosophila or in yeast also prevent recombination (a few species appear to lack this strict dependence, however). The system for generating the DSBs that initiate recombination is generally conserved. Spo11 homologs have been identified in several higher eukaryotes, and a mutation in the Drosophila gene blocks all meiotic recombination.
A few systems are available in which it is possible to compare molecular and cytological events at recombination, but recently there has been progress in analyzing meiosis in Saccharomyces cerevisiae. The relative timing of events is summarized in FIGURE 1.
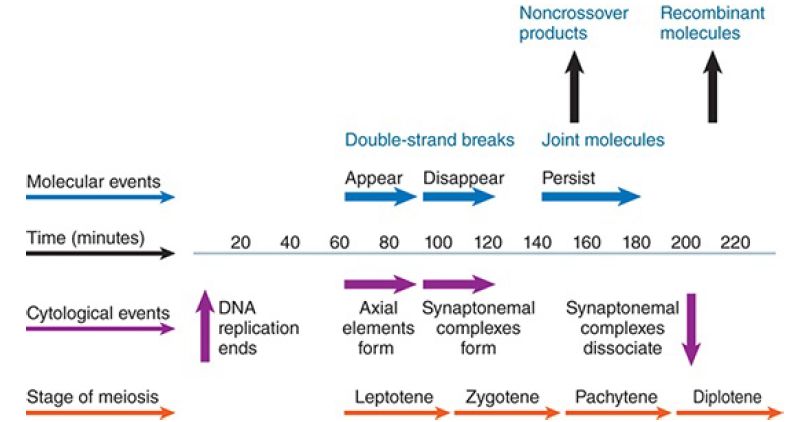
FIGURE 1. Double-strand breaks appear when axial elements form and disappear during the extension of synaptonemal complexes. Joint molecules appear and persist until DNA recombinants are detected at the end of pachytene.
DSBs appear and then disappear over a 60-minute period. The first joint molecules, which are putative recombination intermediates, appear soon after the DSBs disappear. The sequence of events suggests that DSBs, individual pairing reactions, and formation of recombinant structures occur in succession at the same chromosomal site.
DSBs appear during the period when axial elements form. They disappear during the conversion of the paired chromosomes into synaptonemal complexes. This relative timing of events suggests that formation of the synaptonemal complex results from the initiation of recombination via the introduction of DSBs and their conversion into later intermediates of recombination. This idea is supported by the observation that the rad50 mutant cannot convert axial elements into synaptonemal complexes. This refutes the traditional view of meiosis that the synaptonemal complex represents the need for chromosome pairing to precede the molecular events of recombination.
It has been difficult to determine whether recombination occurs at the stage of synapsis, because recombination is assessed by the appearance of recombinants after the completion of meiosis. By assessing the appearance of recombinants in yeast directly in terms of the production of DNA molecules containing diagnostic restriction sites, though, it has been possible to show that recombinants appear at the end of pachytene. This clearly places the completion of the recombination event after the formation of synaptonemal complexes.
Thus, the synaptonemal complex forms after the DSBs that initiate recombination, and it persists until the formation of recombinant molecules. It does not appear to be necessary for recombination as such, because some mutants that lack a normal synaptonemal complex can generate recombinants. Mutations that abolish recombination, however, also fail to develop a synaptonemal complex. This suggests that the synaptonemal complex forms as a consequence of recombination, following chromosome pairing, and is required for later stages of meiosis.
The DSBR model proposes that resolution of Holliday junctions gives rise to either noncrossover products (with a residual stretch of hybrid DNA) or to crossovers (recombinants), depending on which strands are involved in resolution . Recent measurements of the times of production of noncrossover and crossover molecules, however, suggest that this may not be true.
Crossovers do not appear until well after the first appearance of joint molecules, whereas noncrossovers appear almost simultaneously with the joint molecules (see Figure 1). The appearance of these two types of products corresponds to what is considered two independent phases of meiotic recombination. In the first phase, DSBs are repaired through a SDSA reaction, leading to noncrossover products, whereas in the second phase the DSBR pathway is predominant and results largely in crossover products. The molecular outcomes of these phases are illustrated in FIGURE 2. If both types of product were produced by the same resolution process, however, we would expect them to appear at the same time. The discrepancy in timing suggests that crossovers are produced as previously thought—by resolution of joint molecules—but that other routes, such as SDSA, lead to production of noncrossovers. Current research has uncovered roles for a group of proteins known as ZMMs, which in yeast include the proteins Zip1-4, Msh4 and Msh5 (mismatch repair proteins), Mer3, and Spo16. These proteins are well conserved, include a numberof distinct functions, and have roles in crossover determination, synapsis, and other aspects of recombination.
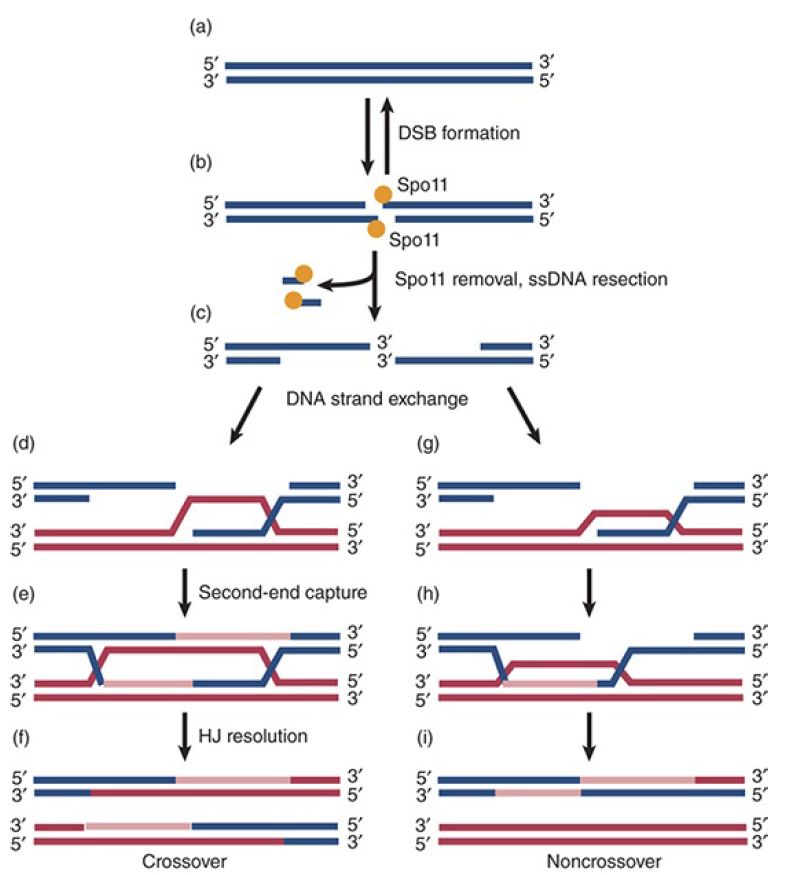
FIGURE 2. Model of meiotic homologous recombination. A DNA duplex (a) is cleaved by Spo11 to form a double-strand break with Spo11 covalently attached to the ends (b). After Spo11 is removed the ends are resected by the MRX/N complex to give single-strand tails with 3′–OH ends, which are complexed with Rad51 and Dmc1.Strand exchange occurs by strand invasion (d and g). Second-end capture results in a double Holliday junction, which is resolved to form crossover products (e and f). Most of the double-strand breaks do not engage in a second-end capture mechanism and instead engage in a synthesis-dependent strand-annealing mechanism (h and i), which results in noncrossover products. Data from M. J. Neale and S. Keeney, Nature 442 (2006): 153–158.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام) قسم الشؤون الفكرية يصدر مجموعة قصصية بعنوان (قلوب بلا مأوى)
قسم الشؤون الفكرية يصدر مجموعة قصصية بعنوان (قلوب بلا مأوى) قسم الشؤون الفكرية يصدر مجموعة قصصية بعنوان (قلوب بلا مأوى)
قسم الشؤون الفكرية يصدر مجموعة قصصية بعنوان (قلوب بلا مأوى)

















