

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Separate Eukaryotic DNA Polymerases Undertake Initiation and Elongation
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
6-4-2021
3840
Separate Eukaryotic DNA Polymerases Undertake Initiation and Elongation
KEY CONCEPTS
- A replication fork has one complex of DNA polymerase α/primase, one complex of DNA polymerase δ, and one complex of DNA polymerase ε.
- The DNA polymerase α/primase complex initiates the synthesis of both DNA strands.
- DNA polymerase ε elongates the leading strand and a second DNA polymerase δ elongates the lagging strand.
Eukaryotic replication is similar in most aspects to bacterial replication. It is semiconservative, bidirectional, and semidiscontinuous. As a result of the greater amount of DNA in a eukaryote, the genome has multiple replicons. Replication takes place during S phase of the cell cycle. Replicons in euchromatin initiate before replicons in heterochromatin; replicons near active genes initiate before replicons near inactive genes. Origins of replication in eukaryotes are not well defined, except for those in yeast (called autonomously replicating sequences [ARS], in S.
cerevisiae). The number of replicons used in any one cycle is tightly controlled. During rapid embryonic development more are activated than in slower-growing adult cells.
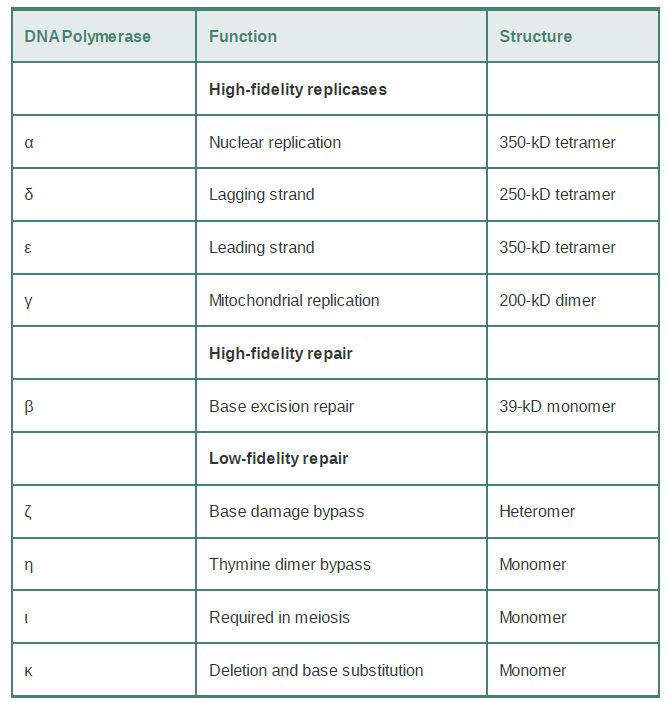
Eukaryotes have a much larger number of DNA polymerases. They can be broadly divided into those required for replication, and repair polymerases involved in repairing damaged DNA. Nuclear DNA replication requires DNA polymerases α, β, and ε. All the other nuclear DNA polymerases are concerned with synthesizing stretches of new DNA to replace damaged material or using damaged DNA as a template. TABLE1 shows that most of the nuclear replicases are large heterotetrameric enzymes. In each case, one of the subunits has the responsibility for catalysis, and the others are concerned with ancillary functions, such as priming or processivity. These enzymes all replicate DNA with high fidelity, as does the slightly less complex mitochondrial enzyme. The repair polymerases have much simpler structures, which often consist ofa single monomeric s ubunit (although it might function in the context of a complex of other repair enzymes). Of the enzymes involved in repair, DNA polymerase β has an intermediate fidelity; all of the others have much greater error rates and are called error-prone polymerases. All mitochondrial DNA replication and recombination is undertaken by DNA polymerase γ.
TABLE 1. Eukaryotic cells have many DNA polymerases. The replication enzymes operate with high fidelity. Except for t he β enzyme, the repair enzymes all have low fidelity. Replicationenzymes have large structures, with separate subunits for different activities. Repair enzymes have much simpler structures.
Each of the three nuclear DNA replication polymerases has a different function, as summarized in TABLE 2.
-DNA polymerase α/primase initiates the synthesis of new strands.
-DNA polymerase ε then elongates the leading strand.
-DNA polymerase δ then elongates the lagging strand.
TABLE 11.3 Similar functions are required at all replication forks.

DNA polymerase α is unusual because it has the ability to initiate a new strand. It is used to initiate both the leading and lagging strands. The enzyme exists as a complex consisting of a 180-kD catalytic (DNA polymerase) subunit, which is associated with three other subunits: the B subunit that appears necessary for assembly, and two small subunits that provide the primase (RNA polymerase) activity. Reflecting its dual capacity to prime and extend chains, this complex is often called pol α/primase.
FIGURE 1 shows that the pol α/primase enzyme binds to the initiation complex at the origin and synthesizes a short strand consisting of approximately10 bases of RNA followed by 20 to 30 bases of DNA (sometimes called iDNA). It is then replaced by an enzyme that will extend the chain. On the leading strand, this isDNA polymera se ε; on the lagging strand this is DNA polymerase δ.
This event is called the polymerase switch. It involves interactions among several components of the initiation complex.
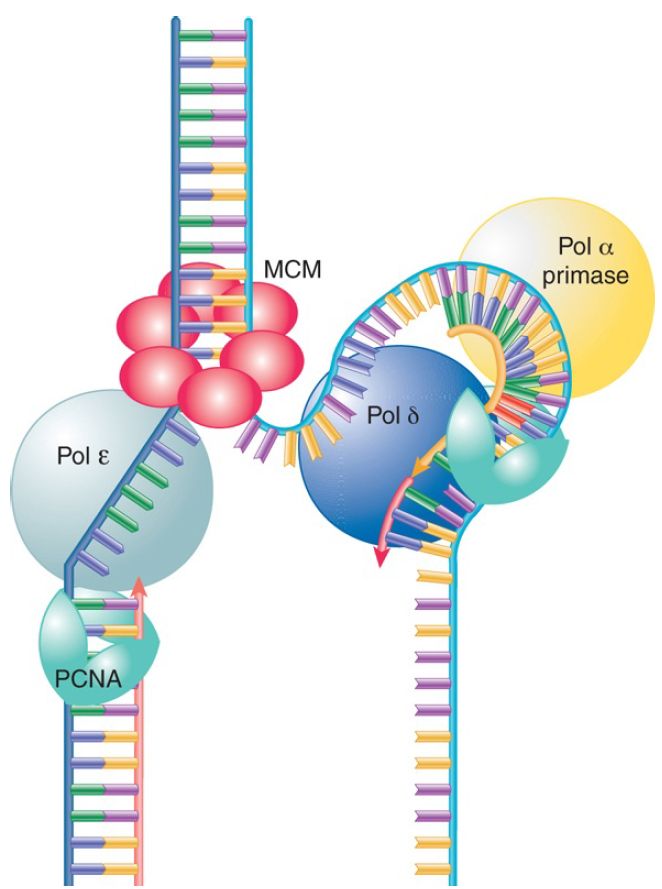
FIGURE 1. Three different DNA polymerases make up the eukaryotic replication fork. Pol α/primase is responsible for primer synthesis on the lagging strand. The MCM helicase (the eukaryotichomolog of Dn aB) unwinds the dsDNA, while PCNA (homolog of α) endows the complex with processivity.
DNA polymerase ε is a highly processive enzyme that continuously synthesizes the leading strand. Its processivity results from its interaction with two other proteins, RFC clamp loader and trimeric PCNA processivity clamp (PCNA was named proliferating cell nuclear antigen for historical reasons).
Table 2 illustrates the conserved function of the replication components extends to the clamp loader and processivity clamp as well other functions of the replisome. The roles of RFC and PCNA are analogous to the E. coli γ clamp loader and β2 processivity unit. RFC is a clamp loader that catalyzes the loading of PCNA onto DNA. It binds to the 3′ end of the DNA and uses ATP hydrolysis to open the ring of PCNA so that it can encircle the DNA. The processivity of DNA polymerase δ is maintained by PCNA, which tethers DNA polymerase δ to the template. The crystal structure of PCNA closely resembles the E.coli β subunit: A trimer forms a ring that surrounds the DNA. The sequence and subunit organization are different from the dimeric β2 clamp; however, the function is likely to be similar.
DNA polymerase α elongates the lagging strand. Like DNA polymerase ε on the leading strand, DNA polymerase δ forms a processive complex with the PCNA clamp. The exonuclease FEN1 removes the RNA primers of Okazaki fragments. The complex of DNA polymerase δ and FEN1 carries out the same type of nick translation that E. coli DNA polymerase I carries out duringOkazaki fragm ent maturation. The enzyme DNA ligase I is specifically required to seal the nicks between the completed Okazaki fragments. Currently, it is not known what factor takes on the function of the E. coli τ dimer that dimerizes thepolymerase complexes in order to ensure coordinated DNA replication.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















