

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Okazaki Fragments Are Linked by Ligase
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
6-4-2021
3230
Okazaki Fragments Are Linked by Ligase
KEY CONCEPTS
- Each Okazaki fragment begins with a primer and stops before the next fragment.
- DNA polymerase I removes the primer and replaces it with DNA.
- DNA ligase makes the bond that connects the 3′ end of one Okazaki fragment to the 5′ beginning of the next fragment.
Researchers can now expand their view of the actions involved in joining Okazaki fragments, as illustrated in FIGURE 1. The complete order of events is uncertain, but it must involve synthesis of RNA primer, its extension with DNA, removal of the RNA primer, its replacement by a stretch of DNA, and the covalent linking of adjacent Okazaki fragments.
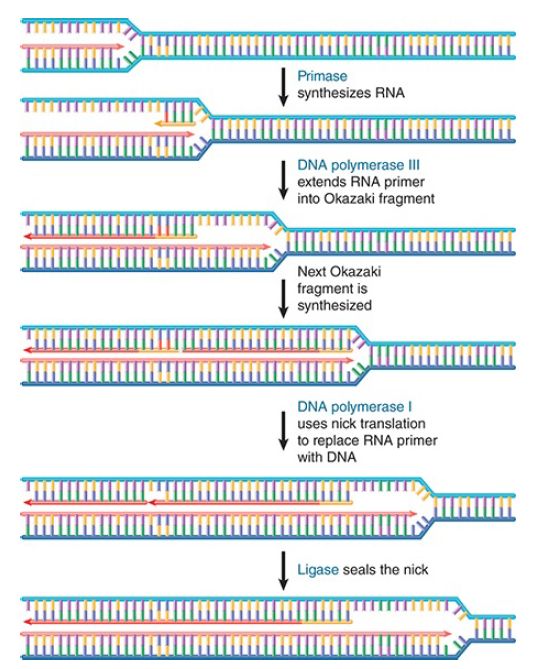
FIGURE 1. Synthesis of Okazaki fragments requires priming, extension, removal of RNA primer, gap filling, and nick ligation.
Synthesis of an Okazaki fragment terminates just before the beginning of the RNA primer of the preceding fragment. When the primer is removed, there will be a gap. The gap is filled by DNA polymerase I; polA mutants fail to join their Okazaki fragments properly. The 5′–3′ exonuclease activity removes the RNA primer while simultaneously replacing it with a DNA sequence extended from the 3′–OH end of the next Okazaki fragment. This is equivalent to nick translation, except that the new DNA replaces a stretch of RNA rather than a segment of DNA.
In mammalian systems (where the DNA polymerase does not have a 5′–3′ exonuclease activity), Okazaki fragments are connected by a two-step process. Synthesis of an Okazaki fragment displaces the RNA primer of the preceding fragment in the form of a “flap.” FIGURE 2 shows that the base of the flap is cleaved by the enzyme FEN1 (flap endonuclease 1). In this reaction, FEN1 functions as an endonuclease, but it also has a 5′–3′ exonuclease activity. In DNA repair reactions, FEN1 can cleave next to a displaced nucleotide and then use its exonuclease activity to remove adjacent material.
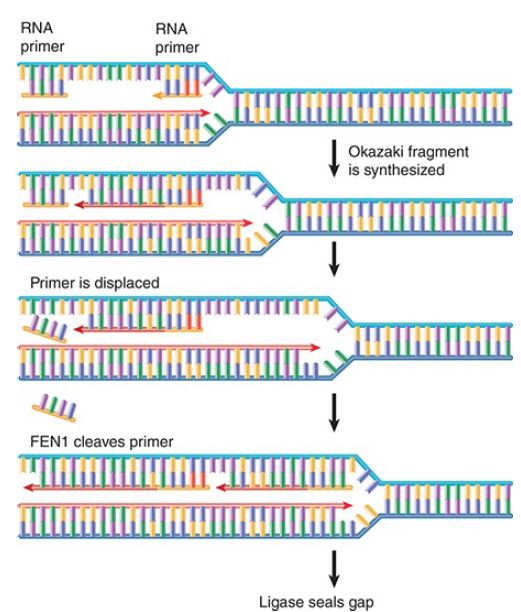
FIGURE 2. FEN1 is an exo-/endonuclease that recognizes the structure created when one strand of DNA is displaced from a duplex as a “flap.” In replication it cleaves at the base of the flap to remove the RNA primer.
Failure to remove a flap rapidly can have important consequences in regions of repeated sequences. Direct repeats can be displaced and misaligned with the template; palindromic sequences can form hairpins. These structures can change the number of repeats . The general importanceof FEN1 is th at it prevents flaps of DNA from generating structures that can cause deletions or duplications in the genome.
After the RNA has been removed and replaced, the adjacent Okazaki fragments must be linked together. The 3′–OH end of one fragment is adjacent to the 5′–phosphate end of the previous fragment. The enzyme DNA ligase makes a bond by using a complex with AMP. FIGURE 3. shows that the AMP of the enzyme complex becomes attached to the 5′ phosphate of the nick and then a phosphodiester bond is formed with the 3′–OH terminus of the nick, releasing the enzyme and the AMP. Ligases are present in both prokaryotes and eukaryotes.
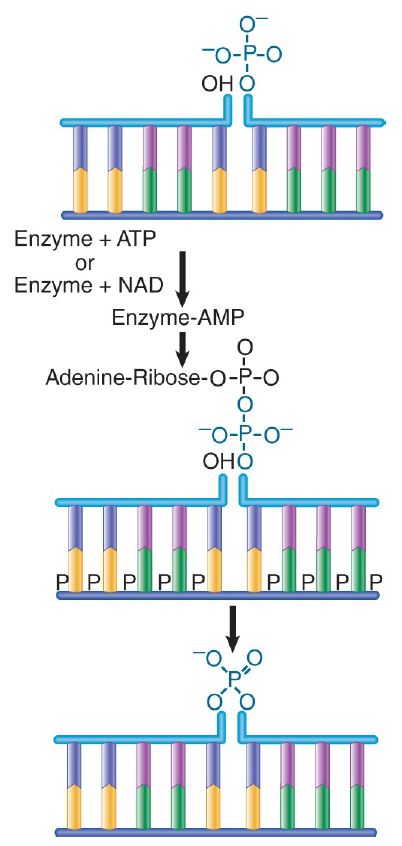
FIGURE 3. DNA ligase seals nicks between adjacent nucleotides by employing an enzyme–AMP intermediate.
The E. coli and Φ T4 ligases share the property of sealing nicks that have 3′–OH and 5′–phosphate termini, as illustrated in Figure 3. Both enzymes undertake a two-step reaction that involves an enzyme–AMP complex. (The E. coli and T4 enzymes use different cofactors. The E. coli enzyme uses nicotinamide adenine dinucleotide [NAD] as a cofactor, whereas the T4 enzyme uses ATP.) The AMP of the enzyme complex becomes attached to the 5′ phosphate of the nick, and then a phosphodiester bond is formed with the 3′–OH terminus of the nick, releasing the enzyme and theAMP.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















