
Replication Requires a Helicase and a Single-Stranded Binding Protein
 المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
 المصدر:
LEWIN’S GENES XII
المصدر:
LEWIN’S GENES XII
 الجزء والصفحة:
الجزء والصفحة:
 5-4-2021
5-4-2021
 1951
1951
Replication Requires a Helicase and a Single-Stranded Binding Protein
KEY CONCEPTS
- Replication requires a helicase to separate the strands of DNA using energy provided by hydrolysis of ATP.
- A single-stranded DNA-binding protein is required to maintain the separated strands.
As the replication fork advances, it unwinds the duplex DNA. One of the template strands is rapidly converted to duplex DNA as the leading daughter strand is synthesized. The other remains single stranded until a sufficient length has been exposed to initiate synthesis of an Okazaki fragment complementary to the lagging strand in the backward direction. The generation and maintenance of single-stranded DNA is therefore a crucial aspect of replication.
Two types of function are needed to convert double-stranded DNA to the single-stranded state:
- A helicase is an enzyme that separates (or melts) the strands of DNA, usually using the hydrolysis of ATP to provide the necessary energy.
- A single-stranded binding protein (SSB) binds to the singlestranded DNA, protecting it and preventing it from reforming the duplex state. The SSB binds typically in a cooperative manner in which the binding of additional monomers to the existing complex is enhanced. The E. coli SSB is a tetramer; eukaryotic SSB (also known as RPA) is a trimer.
Helicases separate the strands of a duplex nucleic acid in a variety of situations, ranging from strand separation at the growing point of a replication fork to catalyzing migration of Holliday (recombination) junctions along DNA. There are 12 different helicases in E. coli. A helicase is generally multimeric. A common form of helicase is a hexamer. This typically translocates along DNA by using its multimeric structure to provide multiple DNA-binding sites.
FIGURE 1. shows a generalized schematic model for the action of a hexameric helicase. It is likely to have one conformation that binds to duplex DNA and another that binds to single-stranded DNA. Alternation between them drives the motor that melts the duplex and requires ATP hydrolysis—typically 1 ATP is hydrolyzedfor each bp that is unwound. A helicase usually initiates unwinding at a single-stranded region adjacent to a duplex. Note that it cannot unwind a segment of duplex DNA; it can only continue to unwind a sequence that has been started . It might function with a particular polarity, preferring single-stranded DNA with a 3′ end (3′–5′ helicase) or with a 5′ end (5′–3′ helicase). A 5′–3′ helicase is shown in Figure 1. Hexameric helicases typically encircle the DNA, which allows them to unwind DNA processively for many kilobases. This property makes them ideally suited as replicative DNA helicases.
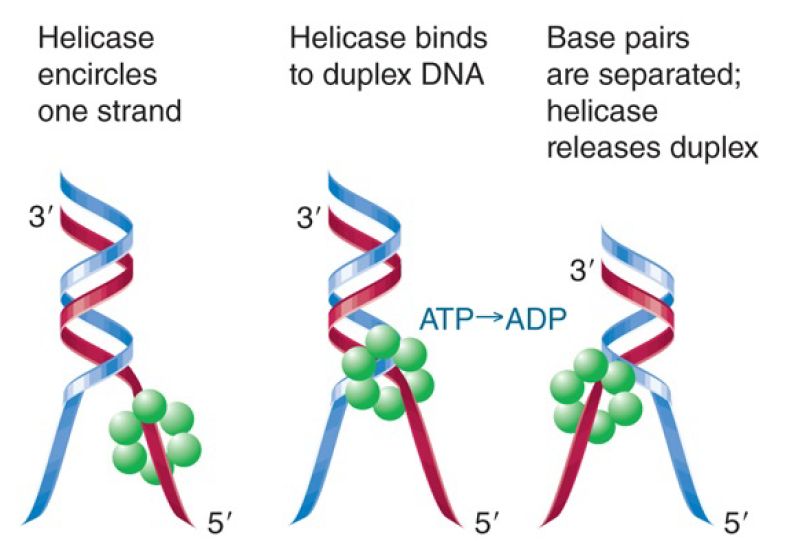
FIGURE 1. A hexameric helicase moves along one strand of DNA. It probably changes conformation when it binds to the duplex, uses ATP hydrolysis to separate the strands, and then returns to the conformation it has when bound only to a single strand.
Unwinding of double-stranded DNA by a helicase generates two single strands that are then bound by SSB. E. coli SSB is a tetramer of 74 kD that binds single-stranded DNA cooperatively.
The significance of the cooperative mode of binding is that the binding of one protein molecule makes it much easier for another to bind. Thus, once the binding reaction has started on a particular DNA molecule, it is rapidly extended until all of the single-stranded DNA is covered with the SSB protein. Note that this protein is not a DNA-unwinding protein; its function is to stabilize DNA that is already in the single-stranded condition.
Under normal circumstances in vivo, the unwinding, coating, and replication reactions proceed in tandem. The SSB protein binds to DNA as the replication fork advances, keeping the two parental strands separate so that they are in the appropriate condition to act as templates. SSB protein is needed in stoichiometric amounts at the replication fork. It is required for more than one stage of replication; ssb mutants have a quick-stop phenotype, and are defective in repair and recombination as well as in replication.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


