

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Minisatellites Are Useful for DNA Profiling
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
21-3-2021
2959
Minisatellites Are Useful for DNA Profiling
KEY CONCEPT
Researchers can use the variation between microsatellites or minisatellites in individual genomes to identify heredity unequivocally by showing that 50% of the bands in an individual are inherited from a particular parent.
Sequences that resemble satellites (in that they consist of tandem repeats of a short unit) but that overall are much shorter— consisting of (for example) 5 to 50 repeats—are common in mammalian genomes. They were discovered by chance as fragments whose size is extremely variable in genomic libraries of human DNA. The variability is observed when a population contains fragments of many different sizes that represent the same genomic region; when individuals are examined, there is extensive polymorphism and many different alleles can be found.
Whether a repeat cluster is called a minisatellite or a microsatellite depends on both the length of the repeat unit and the number of repeats in the cluster. The name microsatellite is usually used when the length of the repeating unit is less than 10 bp; the number of repeats is smaller than that of minisatellites. The name minisatellite is used when the length of the repeating unit is roughly 10 to 100 bp and there is a greater number of repeats. However, the terminology is not precisely defined. These types of sequences are also called variable number tandem repeat (VNTR) regions.
VNTRs used in human forensics are microsatellites that generally have fewer than 20 copies of a 2- to 6-bp repeat. The cause of the variation between individual genomes at microsatellites or minisatellites is that individual alleles have different numbers of the repeating unit. For example, one minisatellite has a repeat length of 64 bp and is found in the population with the following approximate distribution:
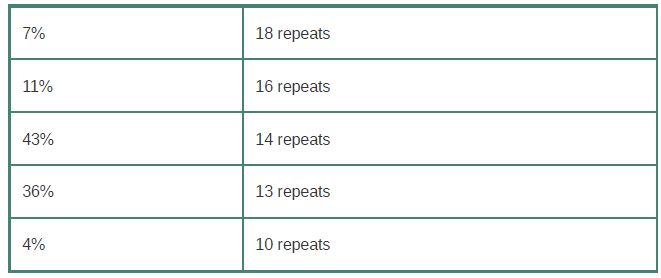
The rate of genetic exchange at minisatellite sequences is high, about 10 per kb of DNA. (The frequency of exchanges per actual locus is assumed to be proportional to the length of the minisatellite.) This rate is about 10-4 times greater than the rate of homologous recombination at meiosis for any random DNA sequence.
The high variability of minisatellites makes them especially useful for DNA profiling, because there is a high probability that individuals will vary in their alleles at such a locus. FIGURE 1 presents an example of mapping by minisatellites. This shows an extreme case in which two individuals are both heterozygous at a minisatellite locus, and in fact all four alleles are different. All progeny gain one allele from each parent in the usual way and it is possible to unambiguously determine the source of every allele in the progeny.
In the terminology of human genetics, the meioses described in this figure are highly informative because of the variation between alleles.
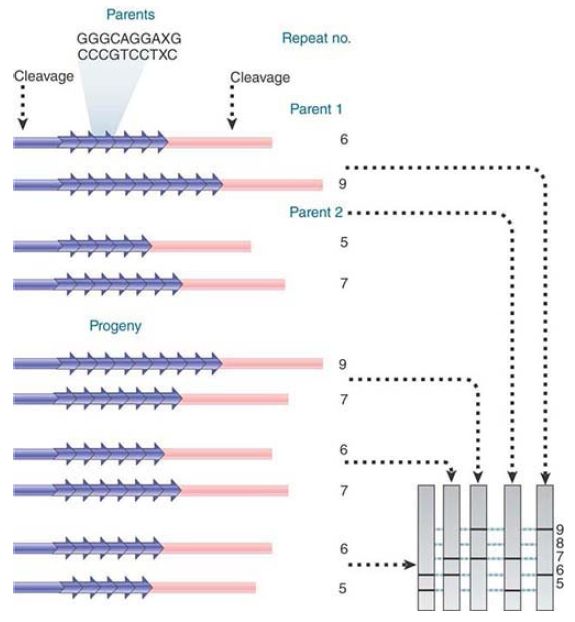
FIGURE .1 Alleles can differ in the number of repeats at a minisatellite locus so that digestion on either side generates restriction fragments that differ in length. By using a minisatellite with alleles that differ between parents, the pattern of inheritance can be followed.
One family of minisatellites in the human genome shares a common “core” sequence. The core is a GC-rich sequence of 10 to 15 bp, showing an asymmetry of purine/pyrimidine distribution on the two strands. Each individual minisatellite has a variant of the core sequence, but about 1,000 minisatellites can be detected on a Southern blot by a probe consisting of the core sequence.
Consider the situation shown in Figure 2 but multiplied many times by the existence of many such sequences. The effect of the variation at individual loci is to create a unique pattern for every individual. This makes it possible to unambiguously assign heredity between parents and progeny by showing that 50% of the bands in any individual are inherited from a particular parent. This is the basis of the technique known as DNA profiling.
Both microsatellites and minisatellites are unstable, although for different reasons. Microsatellites undergo intrastrand mispairing, when slippage during replication leads to expansion of the repeat, as shown in FIGURE 2. Systems that repair damage to DNA—in particular, those that recognize mismatched base pairs—are important in reversing such changes, as shown by a large increase in frequency when repair genes are inactivated. Mutations in repair systems are an important contributory factor in the development of cancer, so tumor cells often display variations in microsatellite sequences.
Minisatellites undergo the same sort of unequal crossing-over between repeats that we have discussed for other repeating units.
One telling case is that increased variation is associated with a recombination hotspot. The recombination event is not usually associated with recombination between flanking markers but has a complex form in which the new mutant allele gains information from both the sister chromatid and the other (homologous) chromosome.
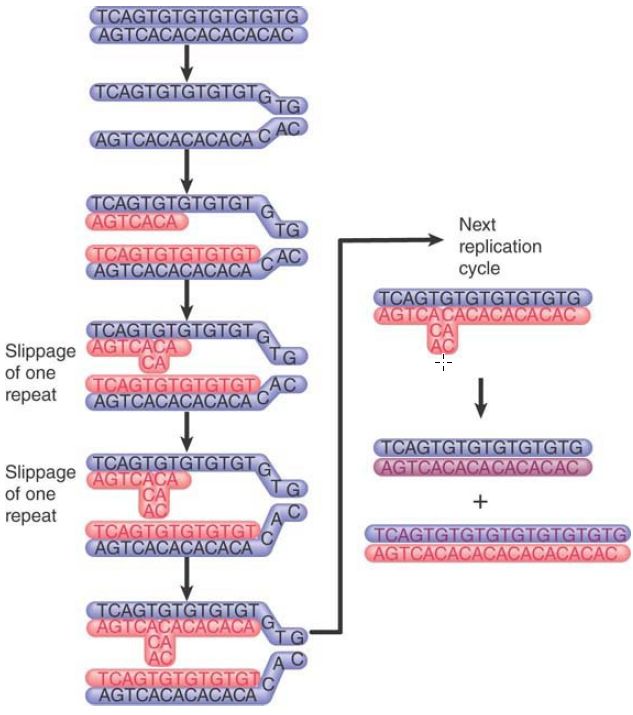
FIGURE 2. Replication slippage occurs when the daughter strand slips back one repeating unit in pairing with the template strand. Each slippage event adds one repeating unit to the daughter strand. The extra repeats are extruded as a single-strand loop. Replication of this daughter strand in the next cycle generates a duplex DNA with an increased number of repeats.
It is not clear at what repeating length the cause of the variation shifts from replication slippage to unequal crossing-over.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















