
Unequal Crossing-Over Rearranges Gene Clusters
 المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
 المصدر:
LEWIN’S GENES XII
المصدر:
LEWIN’S GENES XII
 الجزء والصفحة:
الجزء والصفحة:
 17-3-2021
17-3-2021
 3789
3789
Unequal Crossing-Over Rearranges Gene Clusters
KEY CONCEPTS
-When a genome contains a cluster of genes with related sequences, mispairing between nonallelic loci can cause unequal crossing-over. This produces a deletion in one recombinant chromosome and a corresponding duplication in the other.
-Different thalassemias are caused by various deletions that eliminate α- or β-globin genes. The severity of the disease depends on the individual deletion.
Over a sufficiently long period of time, there are many opportunities for rearrangement in a cluster of related or identical genes. We can see the results by comparing the mammalian α-globin clusters. Although all β-globin clusters serve the same function and have the same general organization, each is different in size, there is variation in the total number and types of β-globin genes, and the numbers and structures of pseudogenes are different. All of these changes must have occurred since the mammalian radiation approximately 85 million years ago (the time of the common ancestor to all the mammals).
The comparison makes the general point that gene duplication, rearrangement, and variation are as important factors in evolution as the slow accumulation of point mutations in individual genes . What types of mechanisms are responsible for gene reorganization?
As described in the introduction, unequal crossing-over can occur as the result of pairing between two sites that are homologous in sequence but not in position. Usually, recombination involves corresponding sequences of DNA held in exact alignment between the two homologous chromosomes. However, when there are two copies of a gene on each chromosome, an occasional misalignment allows pairing between them. (This requires some of the adjacent regions to go unpaired.) This can happen in a region of short repeats or in a gene cluster. FIGURE 1 shows that unequal crossing-over in a gene cluster can have two consequences—quantitative and qualitative:

FIGURE 1. Gene number can be changed by unequal crossingover. If gene 1 of one chromosome pairs with gene 2 of the other chromosome, the other gene copies are excluded from pairing. Recombination between the mispaired genes produces one chromosome with a single (recombinant) copy of the gene and one chromosome with three copies of the gene (one from each parent and one recombinant).
- The number of repeats increases in one chromosome and decreases in the other. In effect, one recombinant chromosome has a deletion and the other has an insertion. This happens regardless of the exact location of the crossover. In the example in Figure 1, the first recombinant has an increase in the number of gene copies from two to three, whereas the second has a decrease from two to one.
- If the recombination event occurs within a gene (as opposed to between genes), the result depends on whether the recombining genes are identical or only related. If the nonhomologous gene copies 1 and 2 are identical in sequence, there is no change in the sequence of either gene. However, unequal crossing-over can also occur when the sequences of adjacent genes are very similar (although the probability is less than when they are identical). In this case, each of the recombinant genes has a sequence that is different from either of the original sequences.
The determination of whether the chromosome has a selective advantage or disadvantage will depend on the consequence of any change in the sequence of the gene product as well as on the
change in the number of gene copies.
An obstacle to unequal crossing-over is presented by the interrupted structure of the genes. In a case such as the globins, the corresponding exons of adjacent gene copies are likely to be similar enough to support pairing; however, the sequences of the introns have diverged appreciably. The restriction of pairing to the exons considerably reduces the continuous length of DNA that can be involved, lowering the chance of unequal crossing-over. So, divergence between introns could enhance the stability of gene clusters by hindering the occurrence of unequal crossing-over.
Thalassemias, inherited blood disorders resulting from abnormal hemoglobin, result from mutations that reduce or prevent synthesis of either α- or β-globin. The occurrence of unequal crossing-over in the human globin gene clusters is revealed by the nature of certain thalassemias. Many of the most severe thalassemias result from deletions of part of a cluster. In at least some cases, the ends of the deletion lie in regions that are homologous, which is exactly what would be expected if it had been generated by unequal crossing-over.
FIGURE 2 summarizes the deletions that cause the α-thalassemias. α-thal-1 deletions are long, varying in the location of the left end, with the positions of the right ends located beyond the known genes. They eliminate both of the α genes. The α-thal-2 deletions are short and eliminate only one of the two α genes. The L deletion removes 4.2 kilobases (kb) of DNA, including the α2 gene. It probably results from unequal crossing-over because the ends of the deletion lie in homologous regions, just to the right of the ψα and α2 genes, respectively. The R deletion results from the removal of exactly 3.7 kb of DNA, the precise distance between the α1 and α2 genes. It appears to have been generated by unequal crossing-over between the α1 and α2 genes themselves.
This is precisely the situation depicted in Figure 1.
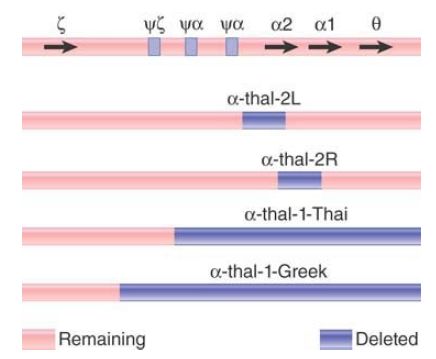
FIGURE 2. α-thalassemias result from various deletions in the α-globin gene cluster.
Depending on the diploid combination of thalassemic alleles, an affected individual can have any number of α chains from zero to three. There are few differences from the wild type (four α genes) in individuals with three or two α genes. However, if an individual has only one α gene, the excess β chains form the unusual tetramer β , which causes hemoglobin H (HbH) disease. The complete absence of α genes results in hydrops fetalis, which is fatal at or before birth.
The same unequal crossing-over that generated the thalassemic chromosome should also have generated a chromosome with three α genes. Individuals with such chromosomes have been identified in several populations. In some populations, the frequency of the triple α locus is about the same as that of the single α locus; in others, the triple α genes are much less common than single α genes. This suggests that (unknown) selective factors operate in different populations to adjust the gene numbers.
Variations in the number of α genes are found relatively frequently,which suggests that unequal crossing-over in the cluster must be fairly common. It occurs more often in the α cluster than in the β cluster, possibly because the introns in α genes are much shorter and therefore present less of an impediment to mispairing between nonhomologous loci.
The deletions that cause β-thalassemias are summarized in FIGURE 3. In some (rare) cases, only the β gene is affected. These have a deletion of 600 bp, extending from the second intron
through the 3′ flanking regions. In the other cases, more than one gene of the cluster is affected. Many of the deletions are very long, extending from the 5′ end indicated on the map for more than 50 kb toward the right.

FIGURE 3 Deletions in the β-globin gene cluster cause several types of thalassemia.
The Hb Lepore type provides the classic evidence that deletion can result from unequal crossing-over between linked genes. The β and δ genes differ by roughly 7% in sequence. Unequal crossingover deletes the material between the genes, thus fusing them together . The fused gene produces a single β-like chain that consists of the N-terminal sequence of δ joined to the Cterminal sequence of β.
Several types of Hb Lepore are known, with the difference between them lying in the point of transition from δ to β sequences. Thus, when the δ and β genes pair for unequal crossing-over, the exact point of recombination determines the position at which the switch from δ to β sequence occurs in the amino acid chain.
The reciprocal of this event has been found in the form of Hb anti- Lepore, which is produced by a gene that has the N-terminal part of β and the C-terminal part of δ. The fusion gene lies between normal δ and β genes. Although heterozygotes for this mutation are phenotypically normal, those that also carry a β deletion in trans show a mild β-thalassemia.
Evidence that unequal crossing-over can occur between more distantly related genes is provided by the identification of Hb Kenya, another fused hemoglobin. This contains the N-terminal sequence of the Aγ gene and the C-terminal sequence of the β gene. The fusion must have resulted from unequal crossing-over between Aγ and β, which differ by about 20% in sequence.
From the differences between the globin gene clusters of various mammals, we see that duplication (usually followed by diversification) has been an important feature in the evolution of each cluster. The human thalassemic deletions demonstrate that unequal crossing-over continues to occur in both globin gene clusters. Each such event generates a duplication as well as a deletion, and researchers must account for the fate of both recombinant loci in the population. Deletions can also occur (in principle) by recombination between homologous sequences lying on the same chromosome. This does not generate a corresponding duplication.
It is difficult to estimate the natural frequency of these events because evolutionary forces rapidly adjust the frequencies of the variant clusters in the population. Generally, a contraction in gene number is likely to be deleterious and selected against. However, in some populations, there might be a balancing advantage that maintains the deleted form at a low frequency. In particular, it might be that both homozygous and heterozygous carriers of a thalassemia deletion show resistance to certain infectious diseases, such as malaria. The form of balancing selection that can maintain such a mutation at a higher incidence is that heterozygotes might not show severe symptoms of thalassemia but benefit from the infectious disease resistance; because both normal and mutant alleles are carried by the heterozygote, selection maintains a “balance” of both alleles. Also, in small populations, genetic drift is likely to play a role in eliminating effectively neutral new duplications; in this mechanism, rare alleles are eliminated from population by chance events. The heterozygote again might not show symptoms, but if heterozygotes are rare in a population, they might either fail to reproduce or happen to not pass along the mutant allele, so the allele is lost from the population.
The structures of the present human clusters show several duplications that attest to the importance of such mechanisms. The functional sequences include two α genes encoding the same polypeptide, fairly similar β and δ genes, and two almost identical γ genes. These comparatively recent independent duplications have persisted in the species, not to mention the more ancient duplications that originally generated the various types of globin genes. Other duplications might have given rise to pseudogenes or have been lost. We expect ongoing duplication and deletion to be a feature of all gene clusters.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


