

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Introduction to the Genome Sequences and Evolution
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
11-3-2021
2711
Introduction to the Genome Sequences and Evolution
Since the first complete organismal genomes were sequenced in 1995, both the speed and range of sequencing have greatly improved. The first genomes to be sequenced were small bacterial genomes of less than 2 megabase (Mb) in size. By 2002, the human genome of about 3,200 Mb had been sequenced. Genomes have now been sequenced from a wide range of organisms, including bacteria, archaeans, yeasts, and other unicellular eukaryotes, plants, and animals, including worms, flies, and mammals.
Perhaps the single most important piece of information provided by a genome sequence is the number of genes.
Mycoplasma genitalium, a free-living parasitic bacterium, has the smallest known genome of any organism, with about only 470 genes. The genomes of free-living bacteria have from 1,700 to 7,500 genes. Archaean genomes have a smaller range of 1,500 to 2,700 genes. The smallest unicellular eukaryotic genomes have about 5,300 genes. Nematode worms and fruit flies have roughly 21,700 and 17,000 genes, respectively. Surprisingly, the number rises only to 20,000 to 25,000 for mammalian genomes.
FIGURE 1 summarizes the minimum number of genes found in six groups of organisms. A cell requires a minimum of about 500 genes, a free-living cell requires about 1,500 genes, a eukaryotic cell requires more than 5,000 genes, a multicellular organism requires more than 10,000 genes, and an organism with a nervous system requires more than 13,000 genes. Many species have more than the minimum number of genes required, so the number
of genes can vary widely, even among closely related species.
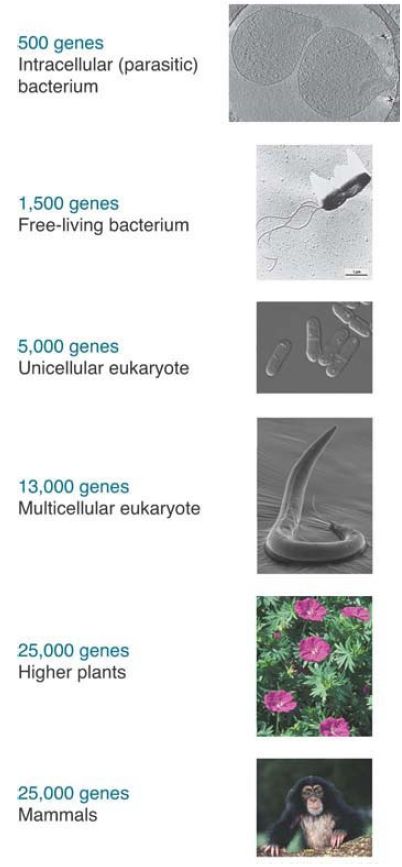
FIGURE .1 The minimum gene number required for any type of organism increases with its complexity.
(a) Photo of intracellular bacterium courtesy of Gregory P. Henderson and Grant J. Jensen,
California Institute of Technology.
(b) Courtesy of Rocky Mountain Laboratories, NIAID, NIH.
(c) Courtesy of Eishi Noguchi, Drexel University College of Medicine.
(d) Courtesy of Carolyn B. Marks and David H. Hall, Albert Einstein College of Medicine,
Bronx, NY.
(e) Courtesy of Keith Weller/USDA.
(f) © Photodisc.
Within prokaryotes and unicellular eukaryotes, most genes are unique. Within multicellular eukaryotic genomes, however, some genes are arranged into families of related members. Of course, some genes are unique (meaning the family has only one member), but many belong to families with 10 or more members. The number of different families may be a better indication of the overall complexity of the organism than the number of genes. Some of the most insightful information comes from comparing genome sequences. The growing number of complete genome sequences has provided valuable opportunities to study genome structure and organization. As genome sequences of related species become available, there are opportunities to compare not only individual gene differences but also large-scale genomic differences in aspects such as gene distribution, the proportions of nonrepetitive and repetitive DNA and their functional potentials, and the number of copies of repetitive sequences. By making these comparisons, we can gain insight into the historical genetic events that have shaped the genomes of individual species and of the adaptive and nonadaptive forces at work following these events.
For example, with the sequences now available for both the human and chimpanzee genomes, it is possible to begin to address some of the questions about what makes humans unique.
The availability of the genome sequences of genetic “model organisms” (e.g., Escherichia coli, yeast, Drosophila, Arabidopsis, and humans) in the late 1990s and early 2000s allowed comparisons between major taxonomic groups such as prokaryote versus eukaryote, animal versus plant, or vertebrate versus invertebrate. More recently, data from multiple genomes within lower-level taxonomic groups (classes down to genera) have allowed closer examination of genome evolution. Such comparisons have the advantage of highlighting changes that have occurred much more recently and are less obscured by additional changes, such as multiple mutations at the same site. In addition,
evolutionary events specific to a taxonomic group can be explored. For example, human–chimpanzee comparisons can provide information about primate-specific genome evolution, particularly when compared with an outgroup (a species that is less closely related, but close enough to show substantial similarity) such as the mouse. One recent milestone in this field of comparative genomics is the completion of genome sequences of nearly 30 species of the genus Drosophila. These types of fine-scale comparisons will continue as more genomes from the same species become available.
What questions can be addressed by comparative genomics? First, the evolution of individual genes can be explored by comparing genes descended from a common ancestor. To some extent, the evolution of a genome is a result of the evolution of a collection of individual genes, so comparisons of homologous sequences within and between genomes can help to answer questions about the adaptive (i.e., naturally selected) and nonadaptive changes that occur to these sequences. The forces that shape coding sequences are usually quite different from those that affect noncoding regions (e.g., introns, untranslated regions, or regulatory regions) of the same gene: Coding and regulatory regions more directly influence phenotype (though in different ways), making selection a more important aspect of their evolution than for noncoding regions. Second, researchers can also explore the mechanisms that result in changes in the structure of the genome, such as gene duplication, expansion and contraction of repetitive arrays, transposition, and polyploidization.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















