

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الحيوية

مواضيع عامة في المضادات الحيوية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
DNA Sequencing
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
7-3-2021
2991
DNA Sequencing
KEY CONCEPTS
-Classic chain termination sequencing uses dideoxynucleotides (ddNTPs) to terminate DNA synthesis at particular nucleotides.
-Fluorescently tagged ddNTPs and capillary gel electrophoresis allow automated, high-throughput DNA sequencing.
-The next generations of sequencing techniques aim to increase automation and decrease time and cost of sequencing.
The classic method of DNA sequencing called dideoxy sequencing has not changed significantly since Frederick Sanger and colleagues developed the technique in 1977. This method requires many identical copies of the DNA, either through cloning or by PCR, an oligonucleotide primer that is complementary to a short stretch of the DNA, DNA polymerase, deoxynucleotides (dNTPS: dATP, dCTP, dGTP, and dTTP), and dideoxynucleotides (ddNTPS). Dideoxynucleotides are modified nucleotides that can be incorporated into the growing DNA strand but lack the 3′ hydroxyl group needed to attach the next nucleotide. Thus, their incorporation terminates the synthesis reaction. The ddNTPs are added at much lower concentrations than the normal nucleotides so that they are incorporated at low rates, randomly.
Originally, four separate reactions were necessary, with a single different ddNTP added to each one. The reason for this was that the strands were labeled with radioisotopes and could not be distinguished from each other on the basis of the label. Thus, the reactions were loaded into adjacent lanes on a denaturing acrylamide gel and separated by electrophoresis at a resolution that distinguished between strands differing by a length of one nucleotide. The gel was transferred to a solid support, dried, and exposed to film. The results were read from top to bottom, with a band appearing in the ddATP lane indicating that the strand terminated with an adenine, the next band appearing in the ddTTP lane indicating that the next base was a thymine, and so on. Read lengths were typically 500 to 1,000 bp.
A major advance was the use of a different fluorescent label for each ddNTP in place of radioactivity. This allowed a single reaction to be run that is read as the strands are hit with a laser and pass by an optical sensor. The information about which ddNTP terminated the fragment is fed directly into a computer. The second modification was the replacement of large slabs of polyacrylamide gels with very thin, long, glass capillary tubes filled with gel (as described previously in the section DNA Separation Techniques).
These tubes can dissipate heat more rapidly, allowing the electrophoresis to be run at a higher voltage, greatly reducing the time required for separation. A schematic illustrating this process is shown in FIGURE 1. As the figure illustrates, the process is automated and machine based. These modifications, with their resulting automation and increased throughput, ushered in the era of whole-genome sequencing. This was the process used to sequence the first set of genomes, including the human genome. It was relatively slow and very expensive. The determination of the human genome sequence took several years and cost several billion dollars to complete.
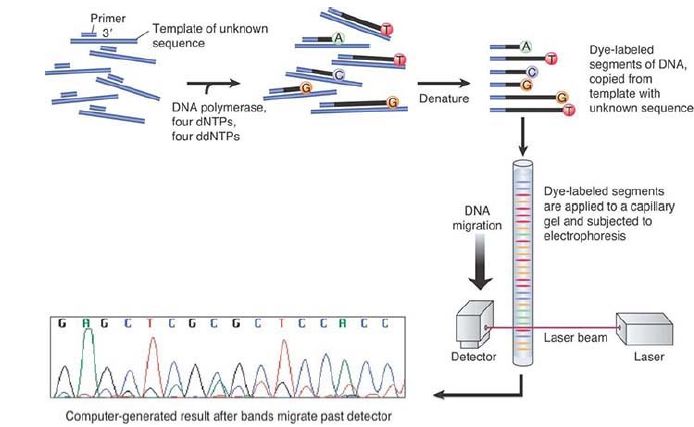
FIGURE 1. DideoxyNTP sequencing using fluorescent tags.
The next generation of sequencing technologies that followed sought to eliminate the need for time-consuming gel separation and reliance on human labor. Modifications of procedures and new instrumentation beginning in about 2005—sometimes called nextgeneration sequencing (NGS) or (now) second-generation NGS— aided in the automation and scaling up of the procedure. This still required PCR amplification of the starting material, which is first randomly fragmented and then amplified. Individual amplified fragments (typically very short—a few hundred bp) are anchored to a solid support and read out one base, in one set of fragments, at a time, in a massively parallel array. These modifications allow sequencing on a very large scale at a much lower cost per kb of DNA than the original first-generation methods.
This technology, sometimes called sequencing-by-synthesis or wash-and-scan sequencing, relies on the detection and identification of each nucleotide as it is added to a growing strand.
In one such application, the primer is tethered to a glass surface and the complementary DNA to be sequenced anneals to the primer. Sequencing proceeds by adding polymerase and fluorescently labeled nucleotides individually, washing away any unused dNTPs. After illuminating with a laser, the nucleotide that has been incorporated into the DNA strand can be detected. Other versions use nucleotides with reversible termination so that only one nucleotide can be incorporated at a time even if there is a stretch of homopolymeric DNA (such as a run of adenines). Still another version, called pyrosequencing, detects the release of pyrophosphate from the newly added base. These secondgeneration systems utilize amplification of material to produce massively parallel analysis runs, but the drawback is that there are typically very short read lengths. The data then require computation to stitch them together into what are called contigs (contiguous sequences).
Technology is now moving from this second generation to a set of third-generation NGS systems. Third-generation sequencing is a collection of methods that avoids the problems of amplification by direct sequencing of the material, DNA or RNA, still giving multiple short (but longer than second-generation sequencing) reads by using single-molecule sequencing (SMS) templates fixed to a surface for sequencing. Again, different companies are proposing different platforms that use different methods to examine the single molecules of DNA. Among these real-time sequencing methods in development are nanopore sequencing and tunneling currents sequencing. The first aims to detect individual nucleotides as a DNA sequence is run through a silicone nanopore, the second, through a channel. Tiny transistors are used to control a current passing through the pore. As a nucleotide passes through, it disturbs the current in a manner unique to its chemical structure. If successful, these technologies have the advantage of reading DNA by simply using electronics, with no chemistry or optical detection required. Nevertheless, there are many kinks to work out of the process before it becomes feasible. Other methods under development include examination by electron microscopy and single-base synthesizing. The accuracy might not be as high as secondgeneration systems, but read lengths are longer, approaching 1,000 bp.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















