

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Single Nucleotide Polymorphisms (SNPs)
المؤلف:
John M Walker and Ralph Rapley
المصدر:
Molecular Biology and Biotechnology 5th Edition
الجزء والصفحة:
21-11-2020
3646
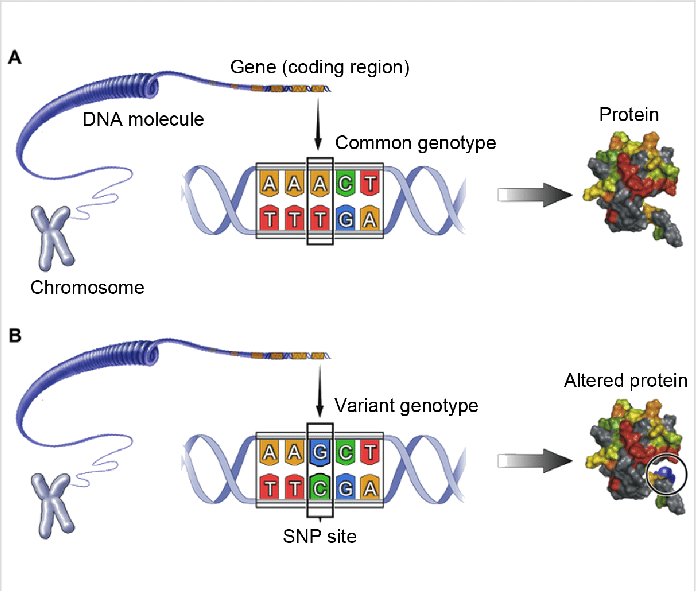
Single Nucleotide Polymorphisms (SNPs)
By far the most widespread type of genetic variants is the single nucleotide polymorphism. Commonly referred to as SNPs (or Snips), these variant sites are found scattered throughout the genomes of most species. An SNP is caused by a single nucleotide replacement such as GAATTC to GTATTC that occurs with a frequency of at least 1% in the population. SNP is a new name for an old phenomenon. SNPs are the cause of most RFLPs and indeed the type of mutation that was indirectly detected in isozyme analysis by zone elctrophoresis before the advent of DNA technology.45 Detailed studies of human DNA suggest that SNPs are extremely abundant: a recent estimate is that there are 10 million SNPs in the human genome,46 which gives an average of one SNP per 300 base pairs. In a recent mapping effort, more than 3 million human SNPs have been analysed in more that 200 individuals. However, SNPs are not distributed evenly throughout the genome and the frequency can vary up to 10-fold.48 Unsurprisingly, because of selection pressure, SNPs are less abundant in the coding regions than non-coding regions; other factors that affect their distribution are local recombination and mutation rates. The majority of SNPs are located in repeat regions which are difficult to analyse; even so, SNPs are the best choice of polymorphic markers that cover the whole genome. Thus it has been estimated that there are 5.3 million common SNPs, each with a frequency of 10–15%, accounting for the bulk of DNA differences. Such SNPs present themselves on average once every 600 base pairs within the human genome in non-coding DNA,50 hence the level of heterozygosity in the human genome resulting from SNPs is extraordinarily high. Many of the current the SNP detection methods can equally detect small insertions and deletions in the DNA. Although SNPs have a use for classical gene mapping by linkage analysis, they have attracted great interest as tools for large-scale association studies with the purpose of associating specific sequences and mutations with specific phenotypes. SNPs can be used simply as genetic markers to identify disease genes by linkage or association analysis, or alternatively the SNPs themselves may on rare occasions be the actual variants in DNA sequences that cause the phenotype. SNPs within genes, especially coding regions that lead to missense mutations, may well change the function, regulation or expression of a protein.52 SNPs have also been discovered in the splice sites of genes and these can result in variant protein products with different exon arrays. A number of SNPs have been discovered in the controlling region of genes and some of these are reported to affect expression and regulation of proteins. It should be emphasised thatmo st SNPs occur in non-coding sequences far away from functional genes are therefore unlikely to have any functional significance, unless by chance they were to affect the configuration of a significant motif such as a remote locus control region. SNP analysis is currently seen as the most likely approach to discovering genes and mutations that contribute to disease. The US National Center for Biotechnology Information (NCBI) curates and maintains an SNP database (dbSNP) which contains sequence details of
several million SNPs, together with short insertions and deletions, from a variety of species. The structural simplicity of SNPs compared with microsatellite renders them ideal for full-scale automation and unambiguous scoring of alleles. However, in contrast with the microsatellite loci, because most SNPs represent single base changes or small insertions or deletions, they can have only two alleles and thus their heterozygosity cannot exceed 0.5, whereas microsatellite in contrast may have a vast number of alleles at individual loci with heterozygosities sometimes approaching 1.0. It
should be noted that the low polymorphism information content ofSNP s compared with microsatellites is more than offset by the fact that SNPs are present in such great abundance throughout the genome.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















