x

هدف البحث
بحث في العناوين
بحث في اسماء الكتب
بحث في اسماء المؤلفين

اختر القسم
موافق


النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الحيوية

مواضيع عامة في المضادات الحيوية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
CpG Islands Are Subject to Methylation
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
12-6-2021
2431
CpG Islands Are Subject to Methylation
KEY CONCEPTS
- Most methyl groups in DNA are found on cytosine on both strands of the CpG doublet.
- Replication converts a fully methylated site to a hemimethylated site.
- Hemimethylated sites are converted to fully methylated sites by a maintenance methyltransferase.
- TET proteins convert 5-methylcytosine to 5- hydroxymethylcytosine to lead to DNA demethylation.
Methylation of DNA occurs at specific sites. In bacteria, it is associated with identifying the bacterial restriction-methylation system used for phage defense and also with distinguishing replicated and nonreplicated DNA. In eukaryotes, its principal known function is connected with the control of transcription:
Methylation of a control region is usually associated with gene inactivation. Methylation in eukaryotes principally occurs at CpG islands in the 5′ regions of some genes; these islands are defined by the presence of an increased density of the dinucleotide sequence CpG .
From 2% to 7% of the cytosines of animal cell DNA are methylated (the value varies with the species). The methylation occurs at the fifth carbon position of cytosine, producing 5-methylcytosine (5mC). Most of the methyl groups are found in CG dinucleotides in CpG islands, where the C residues on both strands of this short palindromic sequence are methylated.
Such a site is described as fully methylated. Consider, though, the consequences of replicating this site. FIGURE 1 shows that each daughter duplex has one methylated strand and one unmethylated strand. Such a site is considered to be hemimethylated.
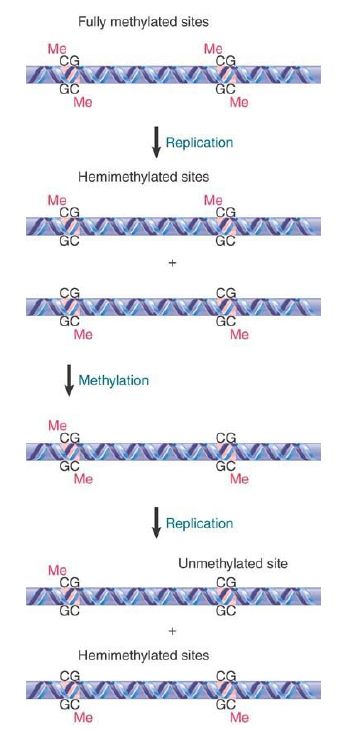
FIGURE 1. The state of methylated CpGs can be perpetuated by an enzyme (Dnmt1) that recognizes only hemimethylated sites as substrates.
The perpetuation of the methylated site now depends on what happens to hemimethylated DNA. If methylation of the unmethylated strand occurs, the site is restored to the fully methylated condition. If replication occurs first, though, the hemimethylated condition will be perpetuated on one daughter duplex, but the site will become unmethylated on the other daughter duplex. FIGURE 2 shows that the state of methylation of DNA is controlled by DNA methyltransferases (often shortened to methylases), or Dnmts, which add methyl groups to the 5 position of cytosine, and demethylases, which remove the methyl groups.
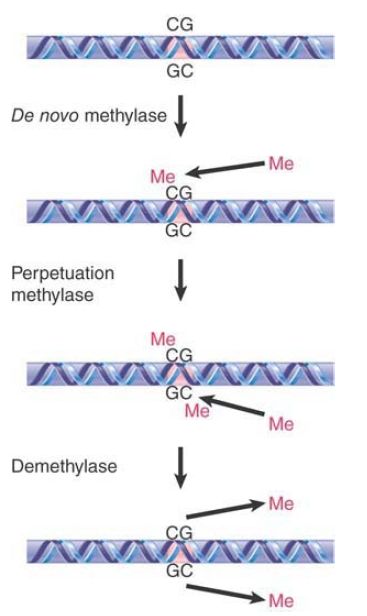
FIGURE 2. The state of methylation is controlled by three types of enzymes. Numerous de novo and perpetuation methylases are known, and methylation occurs in a single enzymatic step. Demethylation is more complex, and no single-step demethylases have been identified.
Two types of DNA methyltransferases have been identified. Their actions are distinguished by the state of the methylated DNA. To modify DNA at a new position requires the action of a de novo methyltransferase, which recognizes DNA by virtue of a specific sequence. It acts only on unmethylated DNA to add a methyl group to one strand. The mouse has two de novo methyltransferases (Dnmt3A and Dnmt3B); they have different target sites, and both are essential for development.
A maintenance methyltransferase acts constitutively only on hemimethylated sites to convert them to fully methylated sites. Its existence means that any methylated site is perpetuated after replication. The mouse has one maintenance methyltransferase (Dnmt1), and it is essential: Mouse embryos in which its gene has been disrupted do not survive past early embryogenesis. Maintenance methylation is almost 100% efficient. The result is that if a de novo methylation occurs on one allele but not on the other the difference will be perpetuated through ensuing cell divisions, maintaining a difference between the alleles that does not depend on their sequences. The fact that maintenance methylation actually falls short of 100% efficiency may lead to a decrease in genomic methylation with progressive cell replication, as is often observed in aging cells. Moreover, this change in methylation status with aging, known as epigenetic drift, is thought to be a contributing factor to the increasing phenotypic variability that is observed with aging of monozygotic twins.
How does a maintenance methyltransferase such as Dnmt1 target methylated CpG sites to preserve DNA methylation patterns with each cell replication? One possibility is that Dnmt1 is brought to hemimethylated sites by factors that recognize methylated CpG sites. Consistent with this concept, a protein has been identified, UHRF1, that is important for the maintenance of methylation both locally and globally through its association with Dnmt1. This protein is able to recognize CpG dinucleotides and to preferentially bind to hemimethylated DNA. Most important, however, is that UHRF1 binds to Dnmt1 and appears to increase the efficacy of Dnmt1 for maintenance methylation at hemimethylated CpG dinucleotides. Thus, UHRF1 has dual functions in recognizing sites for maintenance methylation as well as in recruitment of the
maintenance methyltransferase to these sites for methylation of the unmethylated CpG on the newly synthesized strand, thereby preserving methylation patterns with each cell replication.
Strikingly, UHRF1 also interacts with methylated histone H3, which connects the maintenance of DNA methylation with the stabilization of heterochromatin structure . DNA methylation and heterochromatin are, in fact, mutually reinforcing in several ways, such as in the example depicted in FIGURE 3. Recall that HP1 is recruited to regions in which histone H3 has been methylated at lysine 9, a modification involved in heterochromatin formation. It turns out that HP1 can also interact with Dnmt1, which can promote DNA methylation in the vicinity of HP1 binding. Furthermore, Dnmt1 can directly interact with the methyltransferase responsible for H3K9 methylation, creating a positive feedback loop to ensure continued DNA and histone methylation. These interactions (and other similar networks of interactions) contribute to the stability of epigenetic states, allowing a heterochromatin region to be maintained through many
cell divisions.
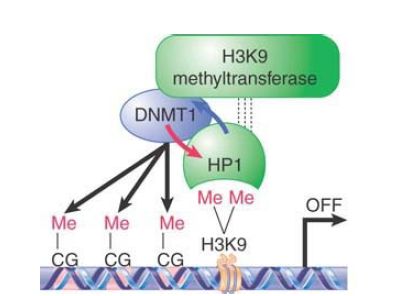
FIGURE 3. Mammalian HP1 is recruited to regions where lysine 9 of histone H3 (H3K9) has been methylated by a histone methyltransferase. HP1 then binds to Dnmt1 and potentiates its DNA methyltransferase activity (blue arrow), thereby enhancing cytosine methylation (meCG) on nearby DNA. Dnmt1 may, in turn, assist HP1 loading onto chromatin (red arrow). Furthermore, association of Dnmt1 with the histone methyltransferase could allow a positive feedback loop to stabilize inactive chromatin.
Methylation has various functional targets. Gene promoters are a common target. The promoter may be methylated when a gene is inactive and is always unmethylated when it is active. The absence of Dnmt1 in mice causes widespread demethylation at promoters; it is assumed that this is lethal because of the uncontrolled gene expression. Satellite DNA is another target. Mutations in Dnmt3B prevent methylation of satellite DNA, which causes centromere instability at the cellular level. Mutations in the corresponding human gene cause the disease ICF (immunodeficiency/centromere instability, facial anomalies). The importance of methylation is emphasized by another human disease, Rett syndrome, which is caused by mutation of the gene encoding the protein MeCP2 that binds methylated CpG sequences. People with Rett syndrome exhibit autism-like symptoms that appear to be the result of a failure of normal gene silencing in the brain.
How are demethylated regions established and maintained? If a DNA site has not been methylated, a protein that recognizes the unmethylated sequence could protect it against methylation. Once a site has been methylated, demethylated sites can be generated in several possible ways. Loss of methylation at a site can occur due to incomplete fidelity of Dnmt1 during maintenance methylation; this is a “passive” demethylation event. Another passive (i.e.,
nonenzymatic) mechanism is to block the maintenance methylase from acting on the site when it is replicated. After a second replication cycle, one of the daughter duplexes will be unmethylated. A third mechanism is to actively demethylate the site, either by removing the methyl group directly from cytosine or by excising the methylated cytosine or cytidine from DNA for replacement by a repair system.
Plants transmit genomic methylation patterns through each generation, though methylation is removed from repeated sequences to prevent interference with nearby gene expression. Plants therefore can easily remove DNA methylation. Plants use the DEMETER family of 5mC DNA glycosylases, followed by cleavage of the DNA backbone phosphodiester bond by apurinic/apyrimidinic (AP) endonuclease and insertion of the unmethylated dCMP base through the base excision repair (BER) pathway .
In mammals, however, the genomic methylation patterns are erased in primordial germ cells—the cells that ultimately give rise to the germline (discussed in the section on imprinting in the chaptertitled Epigenetics II). Primo rdial germ cells have low levels of Dnmt1, thereby eliminating the need for demethylation on larger scales, as seen in plants. This reduced need for DNA demethylation in mammals relative to plants may explain the challenges in characterizing their mechanisms for DNA demethylation. DNMT3A and DNMT3B (de novo methyltransferases) may paradoxically participate in active DNA demethylation in mammals, though. DNMT3A and DNMT3B may possess deaminase activity and are involved in not only gene demethylation but also cyclical demethylation and remethylation within the cell cycle. These enzymes appear to mediate oxidative deamination at cytosine C4 in the absence of the methyl donor (Sadenosylmethionine) to convert 5-methylcytosine to thymine. The resulting guanine-thymine (G-T) mismatch is repaired by base excision, thereby returning the mismatch to a guanine-cytosine (GC) pair and leading to demethylation of a previously methylated CpG site.
Recent work has identified a new family of proteins that may be involved not only in active demethylation but also potentially in producing novel epigenetic marks, such as 5-hydroxymethylcytosine (5hmC). The ten-eleven translocation 1-3, or Tet1-3, proteins are DNA hydroxylase enzymes that can convert 5mC to 5hmC and can further convert 5hmC to 5-formylcytosine(5fC) and then 5-carbox ylcytosine (5caC) in successive reactions These derivatives, especially 5hmC, can be detected in genomic DNA and have been proposed to represent stages of demethylation and to create functionally significant modifications themselves.
Proteins that normally recognize 5mC, such as MeCP2, do not bind to 5hmC, suggesting that generation of 5hmC might serve to reverse methylation-dependent silencing. Similarly, Dnmt1 does not recognize 5hmC during DNA replication, thus the presence of 5hmC can lead to passive demethylation by preventing maintenance methylation. It has also been suggested that, as in plants, 5mC oxidation by TET proteins could also lead to glycosylase action and removal of the methylated site via BER. Alternatively, 5hmC could promote deamination by deaminases such as activation-induced (cytidine) deaminase (AID), which can act on 5mC to create a mismatched T-G base pair or on 5hmC to produce 5-hydroymethyluracil (5hmU), which a repair system can then correct to a standard (unmethylated) C-G pair.
TET proteins/5hmC have been shown to be critical in genome-wide demethylation during zygotic development, and TET proteins also play a role in preventing hematopoietic malignancies (the original identification and name of TET proteins came from the discovery that Tet1 is oncogenically fused to the histone methyltransferase MLL in a translocation in acute myeloid leukemia). Genome-wide analyses in embryonic stem cells have suggested that Tet1 and 5hmC may have important roles in transcriptional regulation. TET proteins (such as Tet1) contain CXXC motifs that bind to CpG islands and may result in maintaining the hypomethylated state of CpG islands at transcriptionally active (or potentially active) sites. Tet1 and 5hmC are enriched at promoters with so-called bivalent domains, which contain histone modifications associated with both active (H3K4me3) and repressive (H3K27me3) states; these types of promoters are usually present in developmentally regulated genes that are poised for expression in particular lineages. Other data suggest that Tet1/5hmC may be involved in both transcriptional activation and repression. Ongoing research is seeking factors that bind to 5hmC or other derivatives to mediate their activities as true epigenetic marks that define the local function of chromatin.